ADBC Summit 2017 Report


Prologue: Many of us in the ADBC world look for ways to expand the community of users of museum collections data and to increase the ways in which collections data are used. Recently, in Trends in Ecology and Evolution (TrEE), an opinion piece was published by Scott A. Morrison, et al. titled "Equipping the 22nd-Century Historical Ecologist." In this paper, Morrison, et al.
Contributed by Pam Soltis and Adania Flemming
iDigBio supported five students in its inaugural mini-REU site program during summer, 2017. This program, modeled on NSF’s Research Experiences for Undergraduates Program, was developed to provide undergraduates with research opportunities using digitized natural history collection data.
Participants in the iDigBio supported Digitizing Mollusks workshop.
The iDigBio supported “Digitizing Mollusks” workshop was held immediately prior to the American Malacological Society meeting in Newark, Delaware on July 15-17, 2017. Thirty-eight collections professionals from 24 established and developing mollusk collections gathered to discuss the status of Mollusk collection digitization in North America and abroad.
Contributed by: Rod Eastwood Curator, Entomological Collection, Eidgenössische Technische Hochschule (ETH) Zürich, Institut für Agrarwissenschaften, Biocommunication & Entomology, Zürich, Switzerland
by Libby Ellwood, Katelin Pearson, Katja Seltmann, Deb Paul and Shelley James
Contributed by Dylan Ricke and Annika Rose-Person, Archbold Biological Station
What is NMITA?

Manta Rays

 From the Darwin Core Hour Team.
From the Darwin Core Hour Team.
Hole-y Plant Databases! Understanding and Preventing Biases in Botanical Big Data
American Horseshoe crab (Limulus polyphemus)

Photo courtesy of: Florida Fish and Wildlife, Photo by: Karen Parker

Contributed by: Teresa Iturriaga, Rhianna Baldree, Alex Kuhn, Andrew Miller

Mycologists long to collect
areas remote to most men
where fungi today may thrive
keeping plants, trees, and cycles alive.
Bridges are to their liking
since one can go underneath
connecting with what lies beneath.
About fungi this is most striking.
Using specimens to create a pollinator community assessment of restored tallgrass prairie
-- Contributed by Heather Cray, Department of Environment and Resource Studies, University of Waterloo

Animal species need space – a place to forage, grow, and nest. This is especially true of Lepidoptera (butterflies and moths), whose caterpillars generally feed exclusively on one genus or species of host plant (think monarch butterflies and milkweed). For the 4,000 or so species of native bees in North America, required forage plants and nesting sites vary from common suburban offerings (e.g., patches of bare ground, maples, willows, clover), to specialized needs which are ecosystem-specific. Enter tallgrass prairie – a grassland ecosystem with high forb diversity that supports a dizzying array of invertebrate life. As our continent’s most endangered ecosystem, the 1-3% that remains is a mix of remnant and restored habitat, and restoration efforts-- both large and small, are ongoing. Read more here.
iDigBio staff members Bruce MacFadden, Libby Ellwood, and Molly Phillips attended the 2017 National Science Teachers Association National Meeting held on March 30-April 2 in the L.A. Convention Center in downtown Los Angeles. The conference was massive – attended by thousands of K-college science teachers from around the country and world.
Publishing a new species? Add the unique identifiers!
Citation of voucher specimen data can be problematic. There are currently no formulated rules for how to cite a digital specimen in a publication, but data aggregators such iDigBio, GBIF, and VertNet offer suggestions. Pensoft is leading the way by providing efficient methods for publishing digital data (see their blog post here) - but it still rarely happens, or occurs in a non-systematic way. Recently, with my colleague Dr George Argent, a new species of Rhododendron from Mount Yule, Papua New Guinea was published in the February 2017 online volume of the Edinburgh Journal of Botany. The digital data for the isotype housed at the Bishop Museum is available through iDigBio and we wanted to cite this information in the published paper. As a test case, we added the Darwin Core occurrenceID and a link to the iDigBio record page. Read more here.

The Society of Herbarium Curators and iDigBio are pleased to announce a 6-week "Strategic Planning for Herbaria” short course.
Take this opportunity to introduce new purpose and excitement into your organization. Learn how to relate your collection’s compelling vision to stakeholders and communicate long-term objectives and strategies to administrators.
Contributed by: Donald H. Pfister, Asa Gray Professor of Systematic Botany and Curator, Farlow Library and Herbarium, Harvard University, 22 Divinity Ave., Cambridge, MA 02138
The scientific view from behind the microphone
Imagine it. The sweaty palms, the nervous fidgeting. You're sitting in the waiting room of the radio station, the governors' office, or waiting to speak with the Chair of your Department. You begin question your preparation - What is the key message and main talking points? Is there an engaging and relevant story to highlight the science? Does it fit with the audience you will be engaging with? You begin cursing that you didn't have more practice!
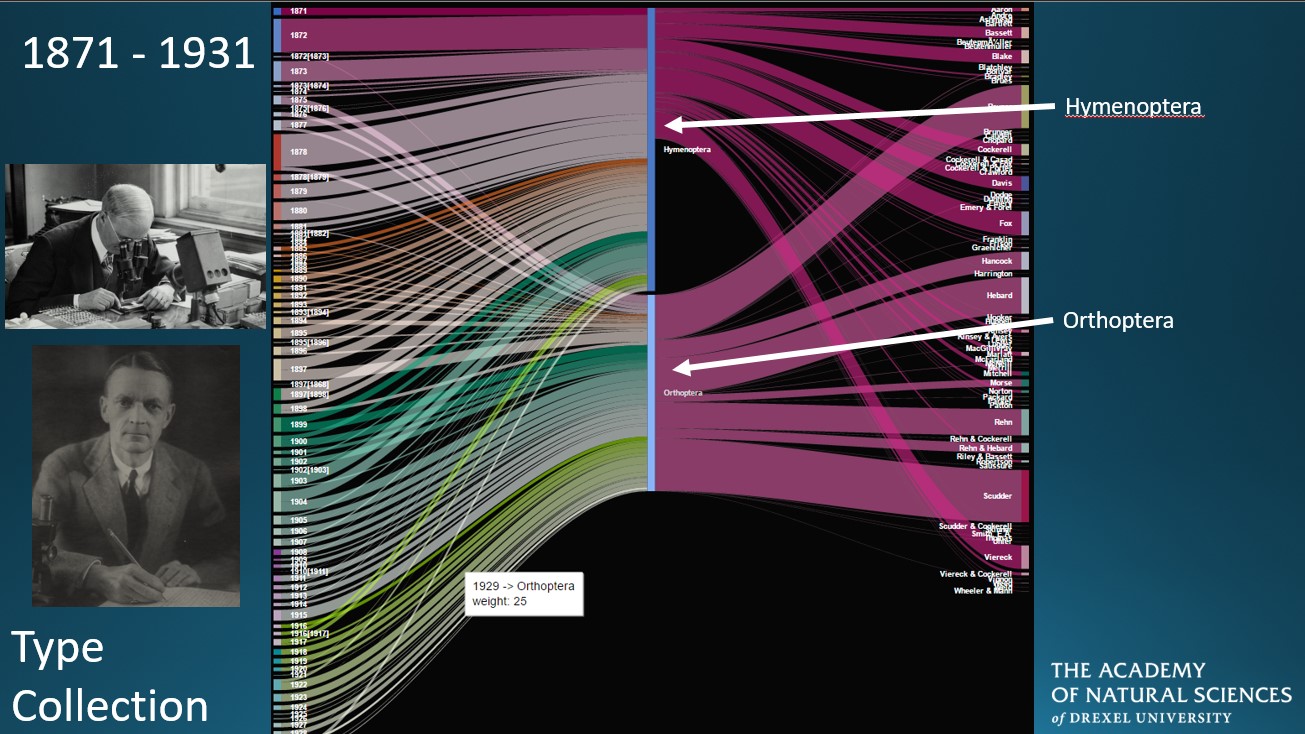
-- Contributed by Vaughn Shirey, The Academy of Natural Sciences of Drexel University
A large portion of my research in The Gelhaus Lab at The Academy of Natural Sciences of Drexel University relies heavily on digitized specimen data and metadata, specifically the who, when, and where of specimen collection. “Big data” research has risen in popularity since high-performance computing has made it easier for researchers to conduct analyses of groups of organisms overnight; however, additional considerations to the use of large datasets should be taken into account. My research focuses on the historical biases present in natural history collection data, including identifying collection bias and gaps in data due to human history. Read more here.
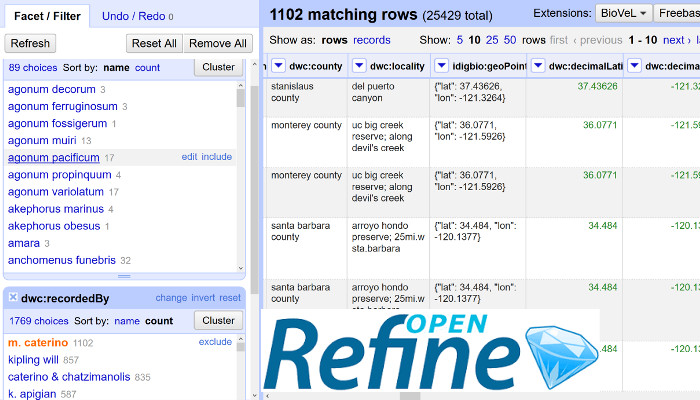
-- Contributed by Chris Evelyn, University of California - Santa Barbara, along with Deborah Paul and Shelley James, iDigBio
This month's Research Spotlight contribution resulted from a recent iDigBio workshop where participants learned the basics of OpenRefine. Finding a limitation to the size of the dataset that could be manipulated, Chris found the following solution to working with large datasets from iDigBio and other biodiversity data aggregators. OpenRefine (formerly Google Refine) is a powerful tool for helping with the cleaning of messy data - ideal for natural history collection managers, data managers, and researchers using biodiversity data alike. Read more here.
Dear iDigBio Enthusiasts,
 -- Contributed by Shelley James & Libby Ellwood, iDigBio
-- Contributed by Shelley James & Libby Ellwood, iDigBio
The Biodiversity Information Standards (TDWG) annual meeting in 2016 had the theme of "Standards Supporting Innovation in Biodiversity and Conservation". Understanding the use of biodiversity standards, and having clear and concise documentation, is essential for the creation, aggregation and downstream use of biodiversity data, and it is exciting to see the diverse TDWG community helping to clarify and expand on the already existing data standards. Read more here.
Contributed by Libby Ellwood and Austin Mast (iDigBio-Florida State University).
