by Deborah Paul, Ana Dal Molin, and Pam Soltis, with contributions from all symposium presenters. Symposium from iDigBio and Universidade Federal do Espírito Santo, Brazil
Overview.
Big Data. There are a few different definitions for what constitutes big data. Most involve enormous datasets. But one could easily argue that research datasets are big when you, as an individual, can no longer work with (analyze, clean, synthesize) said data on your own personal computer. At the 2017 Annual Meeting of the Entomological Society of America Conference (EntSoc), the theme was Ignite. Inspire. Innovate. We set out to do just that with our symposium: Big Data and Bugs: How Massively Collected Biodiversity Data Are Changing the Way We Do Insect Science.
A lot of specimen data now succeeds in getting out of the museum cabinets. These data, along with related biodiversity information, are increasingly available for most scientific inquiry. Over 105 million specimen records are available online to date at iDigBio and are being used to study topics from species distributions and phenology to morphology, conservation, speciation, agriculture, ecology, medicine, and more.
Organizers Ana Dal Molin (Universidade Federal do Espírito Santo, Brazil), Pam Soltis (Florida Museum of Natural History, University of Florida, iDigBio co-PI), and Deborah Paul (Florida State University, iDigBio) invited speakers and also selected talks from the open submissions to foster new collaborations and new voices in our biodiversity data community. The talks covered a range of topics designed to meet interests of all EntSoc member sections: P-IE (Plant–Insect Ecosystems), SysEB (Systematics, Evolution, and Biodiversity), PBT (Physiology, Biochemistry, and Toxicology), and MUVE (Medical, Urban, and Veterinary Entomology).
We were thrilled with attendance: from 75 to 90 people attended our entire session. At the end of the scheduled talks, approximately 20 people participated in a rich discussion that we hope will spur future activities. That discussion gave us the chance to cover even more topics that had not been contemplated in the talks, including the challenges posed by copyright to share information captured from the literature and images, specimen records with locality withheld, and how can data standards and ontologies help integrate information from the literature and ecological observations, besides morphological observations. Some of the suggestions pointed toward continued symposia like this one, along with demonstrations or possibly a workshop on data cleaning, management, and analysis. It was really cool to have entomologists from multiple subdisciplines and career stages participate, from students to senior researchers, museum curators to extension agents. From the discussions, it was also clear that such a mix is atypical in large meetings like this one (with about 3600 attendees!) because one’s tendency is to prioritize talks in a specific area of interest. In a symposium such as this one, we provided the opportunity to see other applications of similar data and to learn that areas that are often seen as “disconnected” or even “rivals” (such as Ecology and Taxonomy) actually face similar challenges when it comes to data management.
Highlights.
Dear reader, you can find all the talks (pdfs) uploaded to the iDigBio Wiki for this Symposium.
Here are some of the big data talk topics:

Linking data. What good are big data if they are hard to synthesize? With more linkages of heterogeneous data, we can better synthesize and use big data to inform science policy, basic research, agriculture, ecology, education, outreach, industry, and to supply inspiration for novel uses. Pam Soltis (iDigBio PI, Florida Museum of Natural History) started our symposium off with the big picture and some great examples of research made possible by linked data. (Try this linked data post if you’re not quite sure what is meant by linked data).
Insect Pest Management (IPM), Agriculture, Economics.
Range mapping. Crystal Klem (Purdue) et al. showed us how collections data inform agriculture about the range of Eudocima phalonia, a fruit-piercing moth. Disagreement about classification and species delineation present management challenges. Current methods of managing E. phalonia (bagging fruit, smoking orchards, hand-netting, pheromone trapping) are not very effective. Detailed, accurate range-mapping does not exist but is needed to support development and implementation of a strategic management plan. With just 30 occurrences per location, meaningful inferences are possible with niche modeling. Crystal digitized and georeferenced a range of specimens, and sampled tissues to sequence DNA to help resolve the systematics issues and produce some range maps. More work to come using MaxEnt, ...
Ecoinformatics, California mandarins, and Scudderia furcata (a katydid). Bodil Cass (UC Davis) revealed her big data approach to studying the damage done to mandarin crops by katydids. She adds collections data to the prior standard observational data. Growers like this because it incorporates their own data into the applied research. The results show that at least for some cases, the IPM guidelines need revisiting as some of the damage is quite minor.
Audience questions:
1. Is a very small percentage of damage (< 1.0%) still of economic import? Yes.
2. Are there databases out there for this agricultural data? Yes.
If your curiosity is piqued, please send Bodil a note!
Citizen science and conservation.
Rare taxa and citizen science. Jaret Daniels (UF McGuire Center) shared how citizen scientists help fill expertise gaps and invertebrate data gaps by gathering much-needed data about rare taxa from sensitive habitats. Using catch and release techniques with an image as digital voucher, 103 trained expert volunteers made over 680 observations. This added knowledge-base helps inform development of effective conservation efforts as such events as hurricanes, human land use, fire suppression, and mosquito abatement all affect biodiversity of invertebrates. Our audience wanted to know if the locality data for these rare taxa were made public. Jaret explained that these data are freely available to the land managers (not directly to the public).
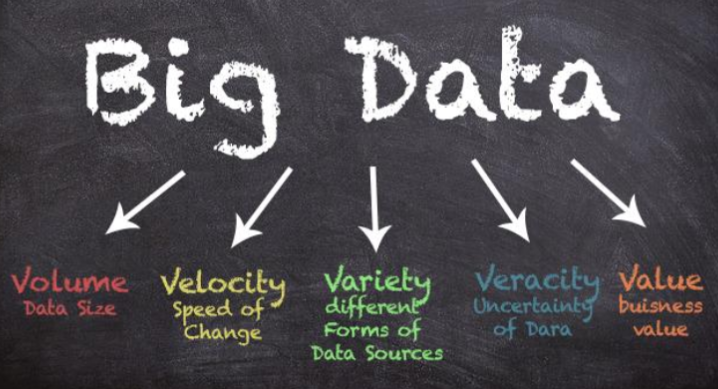
Artificial Intelligence, remote sensing, and citizen science data. Kathleen Prudic (University of Arizona) introduced us to the five “V’s” of big data: Volume (data size), Velocity (speed of change), Variety (different forms of data sources), Veracity (uncertainty of data), and Value (business value). She talked about the power of environmental sensors and citizen scientists to add huge amounts of data to our information about Lepidoptera. In gathering and using these important new data sources, challenges present themselves. Aggregated data often have inconsistent metadata,
many specimens in collections are yet to be digitized, and of course, there is collecting bias. We need past and present data as the past may not predict the present and future accurately. Through many citizen science efforts like iNaturalist and eButterfly we now have a lot more data to look at and discover biodiversity hotspots through species distribution modeling (SDM). But Katy notes there are quality issues. Participants vary in ability to ID organisms, may visit only nearby spots, and have preferences for what they look for. In addition, when taking photos, large lepidoptera get the attention. Experts tend to focus on rare species and mis-ID common as rare; beginners tend to mis-ID rare as common. Artificial intelligence (AI) is being used to take advantage of these new data resources and to address the biases humans bring along with their contributions. And with all these data, Katy encourages us to “start learning R now!”
Patterns of diversity and distribution.
Dead flies and public health. From economic, medical, ecological, and plant-insect viewpoints, we need to know a lot more about Diptera. Erica McAlister provided us with an update on the absurdly large collection of Diptera at the Natural History Museum (NHM) in London. Digitization of over 4 million specimens (the mosquito collection alone is 1.2 million (among pinned specimens, slides, and specimens in alcohol) with 2,400 primary and secondary types, poses unique challenges: with only 2139 individual specimen records digitized at that point, at the current rate of digitization, it would take 150,000 years just to get the mosquito collection digitized (!). But when trying to model mosquito-borne diseases (and other vector-borne diseases), lack of past and present distribution data limits the models (Dr. Steve Le Comber)... and much of this specimen information that has been sitting waiting to be digitized could help to solve this issue, not just for UK mosquitos, but also worldwide. Yikes! So, efforts underway in the NHM Digital Collections Programme include new ways to capture data (and even specimen images) from many slides at once and also to automate data capture from massive samples, like alcohol samples (InSelect), among other ways to speed up sorting. There’s a new workflow that makes it possible to enter specimen data on your phone and have it upload into their Axiell Sapphire database. It was noted yet again that specimen data are biased; however, methods like Dirichlet Process Mixture (DPM) and jack-knifing both show the bias can be accounted for and the data works in the analysis. In addition, new methods are in place (like next-gen-sequencing) to collect and preserve DNA to add to the specimen information available. Look for more work like this soon for the pollinators housed at the NHM.
Swallowtail diversity and distribution. Scientific specimens in collections can offer a robust way to study diversity and distribution. Hannah Owens (University of Florida) et al. investigate climate preference and distribution of swallowtail butterflies in relation to morphological (morphospace) features. After capturing specimen and image data on swallowtails from 4 large insect collections (FLMNH, FM, NMNH, AMNH), the dataset represented 50 / 66 swallowtail species and 1,565 specimen records. Hannah’s paper is in press so I won’t give away what they’ve learned, but you can check out Hannah’s EntSoc talk to learn a bit more!

Historical distributions. From the Field Museum, Crystal Maier introduced us to what’s known about the distribution of Riffle beetles (Elmidae) through the lifelong work of Harry Nelson. A map covered his office walls and ceiling! Uniquely, Harry noted where he did, and did not find riffle beetles – giving present-day researchers a rich source of historical presence and absence data. His field notes need mining but are a bit challenging. Any ideas for how best to tackle this? This dataset offers opportunities for not only distribution studies, but also morphological, ecological, molecular, and who knows what other uses you’ll think up!
Where is that sound coming from? In Lepidoptera, how are sounds produced? What structures make these sounds in the ultrasonic range? David Plotkin (University of Florida) et al. use “nano-CT” with museum specimens to see if they can figure out just how Lepidoptera do this. You might be wondering what nano-CT is, exactly. David gave us a quick tutorial at the beginning of his talk and proceeded to show us how this non-invasive scanning of museum specimens (pixel dimensions of < 1 ) results in detailed 3D reconstructions. Each image can be manipulated to only show selective pieces or layers, giving researchers an elegant way to look at morphology, doing “virtual dissections” (Simonsen 2014). Moths try not to get eaten by the bats and these structures play a role (Kawahara 2015). Take a look at David’s talk to see structures the nano-CT technique has revealed so far!
What is it? Odomatic. Contrasting with Katy’s talk about how artificial intelligence (AI) informs biodiversity research using remote sensing and citizen science data, William Kuhn presented insights into how AI with images taken in nature can be used for automated identification of Odonates. Welcome to Odomatic (under development). To show us just how this works, William gave us a brief introduction to convolutional neural networks. It’s not quite as scary as it sounds – but it is magical. Check out William’s slides to see how we can use the morphological data stored in images we’re amassing as citizen scientists to automatically identify these Odonates and discover new species.
Using collections data to build semantic phenotypes. In this talk by Istvan Miko et al., he shares this definition of big data: “a term for data sets that are so large or complex that traditional data processing application software is inadequate to deal with them.” Istvan shows us how we can use the collections data being aggregated by worldwide collection digitization efforts along with systematically combined transcriptomic data, phylogenomic data, 3D data, character data, anatomy descriptions, and ontologies to construct and model phenotypes.
TaxOnline. Luciane Marinoni is Coordinator of Taxonline – a network of biological collections in Brazil that has been growing quickly and capturing information from small collections hosted at universities, which often don’t get as much attention as the larger museums. From the Universidade Federal do Paraná, Curitiba, Luciane joined us to share information about scientific collections data mobilization in Brazil, specifically in Paraná, and to ask for input on three specific topics:
1. How can we encourage more people to download and use the available data?
2. How can we make this more visible, e. g. for educators, researchers in other areas?
3. How can we identify and convince the stakeholders about the importance of taxonomy and collections?
Great questions, Luciane! There’s lots to discuss on those three topics. Here in the USA, for the ADBC project, all three of those are key challenges everyone is working on, and we look forward to visiting you in Brazil in February 2018 to discuss them in more detail. Readers here may have some insights for you, too.
Posters too. We were pleased to include 2 posters in our session, one titled: Low-cost genomic architecture as a species delimitation tool using rDNA fingerprints by John Sproul, et al. where some of the material analyzed came from museum specimens. The other poster: Students discover: ANTS - Connecting science and education by Daniela Sorger, et al showed us all an intriguing way to get science (and ants!) into the classroom.
Big Insect Collection Dataset Insights. With big (and bigger) data, researchers need strategies to manage and evaluate the data. So much data – so little time! To wrap up our session, Katja Seltmann offered to share her take on “Challenges and trends in really BIG insect collection DATAsets.” In this talk, Katja outlines the steps she takes from downloading a dataset, to getting it research ready. She also discusses the “why” of each step so you can follow her reasoning. She offers direct examples and addresses these issues at the level of the data aggregator, the researcher, and the data provider. Thanks Katja, for tying up all the big data pieces into such a tidy research-ready bundle.
If you’ve made it this far, congratulations!
We hope that we’ve inspired you, and more researchers, to explore the wealth and potential of data in digitized natural history collections.
iDigBio’s Deborah Paul and Kevin Love also took the booth to EntSoc and visited with over 250 entomologists over 3 days. Many signed up for our Newsletter, and we connected with new potential collaborators, data users, and future data providers. In a new twist, entomologists and collection managers joined us in the exhibit to talk with their peers about the importance of collections as well as current and potential education, outreach, and research uses of the data. We did data searches, shared education and outreach materials, and talked about Data Carpentry, too. As ever, everyone got excited when they learned about the Libraries of Life cards – bringing collections to you – in 3D! We’d like to give special thanks to Crystal Maier, William Kuhn, Ana Dal Molin, and others who gave their time and expertise to our ADBC efforts at EntSoc2017.
PS. And if you’re wondering about past events, you can check out our annotated Workshop Summaries list.
References.
Dal Molin A, Paul DL, Soltis P. 2017. Big Data and Bugs: How Massively Collected Biodiversity Data Are Changing the Way We Do Insect Science. Symposium in the SysEB (Systematics, Evolution, and Biodiversity) Section, Annual Meeting of the Entomological Society of America, 7 November 2017.
Kawahara AY, Barber JR. 2015. Tempo and mode of antibat ultrasound production and sonar jamming in the diverse hawkmoth radiation. Proceedings of the National Academy of Sciences (PNAS) 112 (20), 6407-6412 DOI: 10.1073/pnas.1416679112
Simonsen TJ, Kitching IJ. 2014. Virtual dissections through micro-CT scanning: a method for non-destructive genitalia ‘dissections’ of valuable Lepidoptera material. Systematic Entomology 39, 606-618 DOI: 10.1111/syen.12067








