by Heather Appleby, former undergrad intern, Tri-Trophic Thematic Collection Network (TTD-TCN). Katja Seltmann (TTD-TCN), Deb Paul, Alex Thompson, and Matt Collins Eds.
Katja Seltmann (TTD-TCN), Deb Paul, Alex Thompson, and Matt Collins Eds.
Hi Everyone, I'm Heather Appleby. I was an undergraduate intern for the Tri-Trophic Thematic Collection Network (TTD-TCN) for 10 weeks in the summer of 2015. My internship focused on learning the fundamentals of R, while producing a useful product to help the TTD-TCN review georeferenced data for over 1 million specimen records. I endeavored to accomplish these goals in collaboration with iDigBio, by utilizing the iDigBio portal recordset correction feature (https://goo.gl/vvSzDM), while providing both feedback to iDigBio about those error reports, and creating publicly available R-scripts for simplifying the view of the error reports (https://github.com/happleb00/iDigBio_georeference_cleanup).
One of the major objectives of the Tri-Trophic Thematic Collection Network is to make available one million North American Hemiptera specimen records through the iDigBio data portal. At the American Museum of Natural History (AMNH), the principal database used for this effort is Arthropod Easy Capture (AEC; Arthropod Easy Capture, 2014; Schuh, Hewson-Smith, & Ascher, 2010) Database. In total, a group of 50+ paid digitizers and volunteers at 11 institutions used AEC to capture species and collection event information from specimen labels, contributing to a grand total of 902,000 specimen just in the AMNH AEC, with a project wide total reaching 1.3 million records (and still growing).
The specimen databasing procedure for the TTD-TCN is fairly standard. It begins with a group of specimen digitizers, or people who are trained in specimen handling and recording information off of specimens, tasked with capturing information from insect labels and entering it into the AEC database. The labels denote species, collector name, collection number, date, and location, and sometimes latitude and longitude coordinates. If coordinates are not provided on the collection label, the specimen record is reviewed a second time, by different group of digitizers to be georeferenced. Georeferencing, or the process of assigning latitude and longitude coordinates to a collection site locality string, involves searching for this collection location information in GeoLocate web software and Google Earth. Our georeferencing method follows the best practices outlined by iDigBio as well as a "how to" guide specifically designed for the AEC database (see links below). Once coordinates are retrieved, they are entered into the AEC database, along with the previously entered label information for each species and locality. Occasionally records are georeferenced incorrectly during the digitization process. Incorrect georeferences are typically due to human error at some point in the process. Latitude and longitude coordinates may be read incorrectly, localities may be written poorly on the label and, thus, typed incorrectly in the database, or the wrong state, county or municipality may be applied to a locality string.
 Completed specimen records are shared with the iDigBio portal through a Darwin Core Archive. iDigBio detects incorrect locality data, based on latitude and longitude coordinate data and county, state, or country bounds, and proposes corrections to those records. The latest version of the iDigBio portal (released August, 2015) comes with the ability to download their analysis of every provided record set, allowing data providers direct feedback about the record sets they are submitting. This new feature categorizes bad entries and their corrections based on the detected errors, making database cleanup more organized and straightforward.
Completed specimen records are shared with the iDigBio portal through a Darwin Core Archive. iDigBio detects incorrect locality data, based on latitude and longitude coordinate data and county, state, or country bounds, and proposes corrections to those records. The latest version of the iDigBio portal (released August, 2015) comes with the ability to download their analysis of every provided record set, allowing data providers direct feedback about the record sets they are submitting. This new feature categorizes bad entries and their corrections based on the detected errors, making database cleanup more organized and straightforward.
|
iDigBio error flags |
||
|
rev_geocode_eez |
|
The lat/lon point in the ocean, or in a location that is not considered part of an Exclusive Economic Zone, and should be moved inland. |
|
dwc_stateprovince_replaced |
|
The lat/lon does not correspond to the state. Either the coordinates must be re-georeferenced or the state was entered incorrectly. |
|
dwc_country_replaced |
|
The lat/lon does not correspond to the country. Either the coordinates must be re-georeferenced or the country was entered incorrectly. |
|
rev_geocode_mismatch |
|
The latitude and longitude values are switched. The reversed latitude and longitude should be checked in GeoLocate or Google maps and regeoreferenced. |
|
rev_geocode_lon_sign |
|
The longitude sign is flipped. The sign should be switched to negative or positive and checked on GeoLocate or Google Maps and re-georeferenced. |
|
rev_geocode_lat_sign |
|
The latitude sign is flipped. The sign should be switched to negative or positive and checked on GeoLocate or Google Maps and re-georeferenced. |
|
geopoint_similar_coord |
|
The latitude or longitude may have been copied twice, resulting in the same coordinate for both. The locality should be re-georeferenced. |
Each type of flag is packaged up in a downloadable folder. Every folder contains a list of the incorrectly georeferenced data (occurrence_raw.csv) and a list of and the iDigBio corrected records (occurrence.csv). All files are in the Comma Separated Value (CSV) format. The uncorrected file list (occurrence_raw.csv) is useful to search for "bad localities" in our data. Different flags indicate different kinds of errors (see table above). For example, the flag "rev_geocode_eez" indicates the coordinates represent a location in the ocean, not a likely place to collect an insect. Once a bad locality is identified, the incorrect latitude and longitude values are inserted into GeoLocate to verify the error. If it is an error, new coordinates are obtained and the AEC database updated.
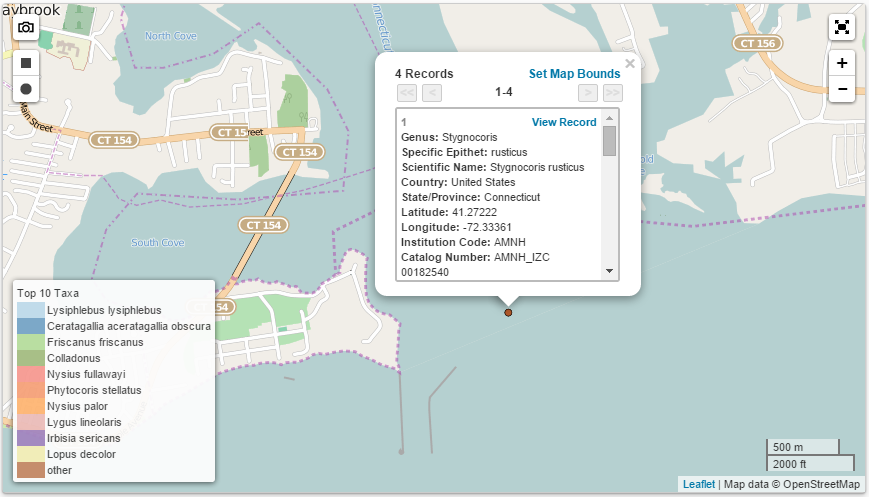
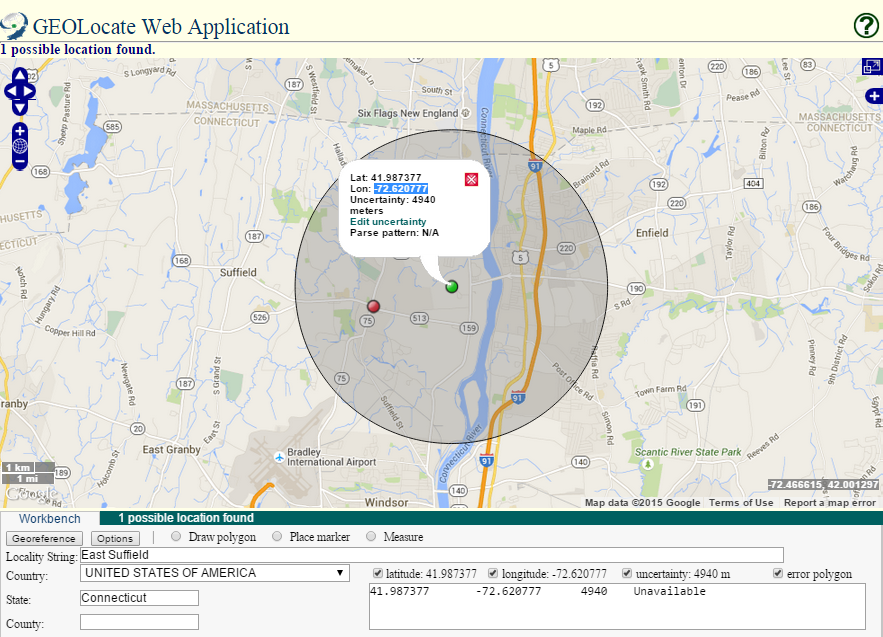
After correcting a few errors, I had come upon a number of duplicate localities in the dataset and some correctly georeferenced entries in the bad locality files. iDigBio files contain all of the Darwin Core fields for all specimens associated with the bad localities, making extraction of only the locality data from these files time consuming. I realized that it would make sense for me to reformat the iDigBio output using R into a list of unique localities with only the locality information fields (i.e. dwc:locality, dwc:county, dwc:decimalLatitude, dwc:decimalLongitude). In addition, I filtered out all localities except US, Mexican, and Canadian localities and made a unique list, removing duplicates. Duplicate localities in the raw files exist because iDigBio outputs specimen data, not specifically locality data, and many specimens may be collected at the same locality. Finally, in an attempt to determine the cause of false positives, I organized the file by country and state, as it appeared that these errors were occurring because the centrad was placed in a different county than what is directly recorded in the Darwin Core county (dwc:county) field directly.
Another objective of this project was to provide feedback for the new recordset evaluation feature in iDigBio. While accessing the iDigBio correction records, and working with the output to fix our incorrectly georeferenced data, I reported several observations to the iDigBio Advanced Computing and Information Systems (ACIS) team. These observations were utilized to help refine the iDigBio correction output. We were able to help identify two bugs in the flagged files through our correspondence. Initially many of the localities showing up in the reverse longitude sign flag appeared to be false positives. When checked in GeoLocate, the coordinates seemed to be very close to a county or state border. Immediately the ACIS team recognized an issue with the designation of boundaries and fixed the problem. We came across another issue while cleaning up the files in the “dwc_stateprovince_replaced” flagged list. All of the georeferenced localities labeled “Baja California Norte” in our database were corrected to “California” in the iDigBio Portal. While “Baja California Norte” is an incorrect state name for the region, we learned that the correction should have been made to “Baja California”.
Note from Katja Seltmann: As a result of Heather’s ten week summer internship, the TTD-TCN project provided useful feedback that helped to guide development of this important community resource. Her introduction to R quickly proved helpful in her current position at the School of Visual Arts NATLab (http://bioart.sva.edu), where she is collecting NYC soil samples and analyzing them for nutrient content (http://2015.igem.org/Team:SVA-NYC).
Note from Deb Paul: The topic of biodiversity Data Quality (DQ) cannot be over-emphasized. How can publishing data with an aggregator like iDigBio promote data quality? The iDigBio Data Management Interest Group (DMI) and Cyberinfrastructure Working Group (CYWG) collaborated with the TTD-TCN to highlight data DQ from the provider and aggregator points-of view. Heather's showing us here how a provider (TTD-TCN) uses the iDigBio DQ flags and coming up in part two of this series, the CYWG explains just how these DQ flags are generated (Webinar: Improving Data Quality: iDigBio Recordset data cleaning method, tools, and data flags, October 23rd, 2015, 2 PM EST).
Important Links:
Project GitHub: https://github.com/happleb00/iDigBio_georeference_cleanup
School of Visual Arts NATLab: http://bioart.sva.edu
TCN Summary: http://tcn.amnh.org/
iDigBio Portal: https://goo.gl/vvSzDM
GeoLocate: http://goo.gl/jizt4G
iDigBio georeferencing practices: http://goo.gl/tbgc4w
Georeferencing how to: https://goo.gl/SZDH45
References:
Arthropod Easy Capture. (2014). Arthropod Easy Capture. Retrieved October 14, 2014, from http://sourceforge.net/projects/arthropodeasy/
Schuh, R. T., Hewson-Smith, S., & Ascher, J. S. (2010). Specimen databases: A case study in entomology using Web-based software. American Entomologist, 56, 206–216.







