Polyploidy in ferns: biodiversity data documenting speciation!
-- Contributed by Blaine Marchant
My research for iDigBio addresses ecological and evolutionary questions by utilizing the enormous dataset provided by digitized natural history specimens from across North America. My current project is aimed at investigating the ecological differentiation of polyploid plant species from their diploid progenitor species. Polyploidy, or whole-genome duplication, has long been recognized as a mechanism for producing reproductive isolation of a population from the parent, or progenitor, species - a critical step for subsequent speciation – especially in plants. However, the means by which the newly formed polyploid species ecologically differs from its progenitor(s) has been highly contended for decades. To tackle this quandary, I am producing ecological niche models (ENMs) using tens of thousands of specimen occurrence records of thirteen polyploid species and their diploid progenitors in order to quantify and assess how these polyploid species ecologically diverge from their parents. Not only will this project answer a question that has interested evolutionary biologists and ecologists for years, but it will also demonstrate the analytical power of digitized specimen data for future researchers.
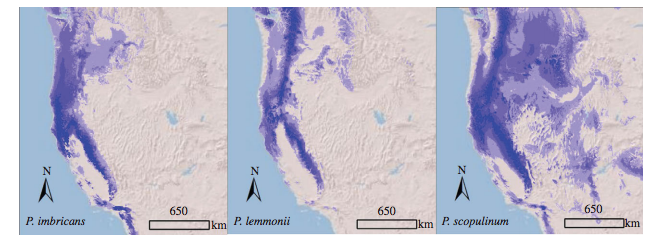
Ecological niche models for the allotetraploid Polystichum scopulinum and its diploid parents, P. imbricans and P. lemmonii, in western North America. Intensity of color indicates probability of occurrence, with lavender designating lower probability than dark blue. From: Soltis, Pamela S., Xiaoxian Liu, D. Blaine Marchant, Clayton J. Visger, and Douglas E. Soltis (2014). "Polyploidy and novelty: Gottlieb's legacy." Philosophical Transactions of the Royal Society of London B: Biological Sciences 369, no. 1648: 20130351.







