Assessment of the pinned specimen digitization progress of the University of Alaska Museum Insect Collection
Ashley L. Smith, Derek S. Sikes, Taylor L. Kane, Adam Haberski, Jayce B. Williamson, Renee K. Nowicki, Michael J. Apperson
University of Alaska Museum, University of Alaska Fairbanks, Fairbanks, Alaska, USA
This article was originally published in the Alaska Entomological Society Newsletter AKES_newsletter_2021_n1_a01.pdf (akentsoc.org)
Introduction
As anthropogenic climate change enlarges our planet’s sixth mass extinction crisis (Ceballos et al. 2015), there is an increasing urgency to museum science (Raven & Miller 2020). Efforts have accelerated to digitize biodiversity data held by the world’s Natural History Collections with a goal of creating open access data according to the FAIR Data Principles: findability, accessibility, interoperability, and reusability (Nelson & Ellis 2019, Heberling et al. 2021). Many museums have a significant number of specimens whose data are not yet being shared online with the scientific community. Such unshared, undigitized data are often called “dark data” (Heidorn 2008, Sikes et al. 2016). Dark data are more difficult to access and are consequently often ignored by biodiversity informaticians using so-called ‘Big Data’ to answer questions of conservation concern. For example, Kerr et al. (2015) analyzed open access, online bumble bee data covering 110 years of collection effort in North America and Europe and detected trends showing range losses and elevational shifts. Specimens with dark data that were relevant to this study were not included because their data were not shared according to FAIR data principles.
The University of Alaska Museum Insect Collection (UAM) is now over two decades old. UAM staff have been able to keep track of and summarize annual growth of the collection’s digitized specimens but had lost track of its undigitized holdings. It is relatively easy to count museum specimen records in a database (Sikes 2015, Whitmore et al. 2020), but specimens that are not yet digitized must be counted manually and doing so only provides a snapshot of the undigitized holdings. As work progresses and new specimens are added to the collection and old undigitized specimens are processed, the count of undigitized specimens becomes inaccurate.
In the spring of 2020, we assessed the digitization progress in the UAM pinned collection. The purpose of the project was to find out how many specimens were undigitized per insect order.
Methods
We started at the first drawer of the pinned insect collection and visually counted any specimens not bearing a UAM barcode label using a hand-held tally counter. All digitized pinned specimens have a Data Matrix barcode label, which sticks out from under the data label and is visible from above (Fig. 1). We proceeded through all the drawers in this manner, grouping counts within each order. We did not count parasites or phoretic specimens such as mites or nematomorphs (eg. UAM:Ento:354713) on pinned insect specimens. A pin with multiple specimens of the same taxon on it (eg. inside a gelatin capsule) was counted as a single undigitized specimen because once digitized it would be a single database record. Our total count is therefore a minimum, not actual, number of pinned specimens (Sikes 2015).

Figure 1. Example of pinned specimens with UAM barcodes visible.
After we finished a first pass at counting, UAM staff used those results to strategically target undigitized specimens for immediate digitization. We therefore present two undigitized counts: the first pass, and the later, most recent counts to show the effectiveness of this strategic approach. We used the most recent counts to calculate the percent digitized for each order and for the entire pinned collection.
Results
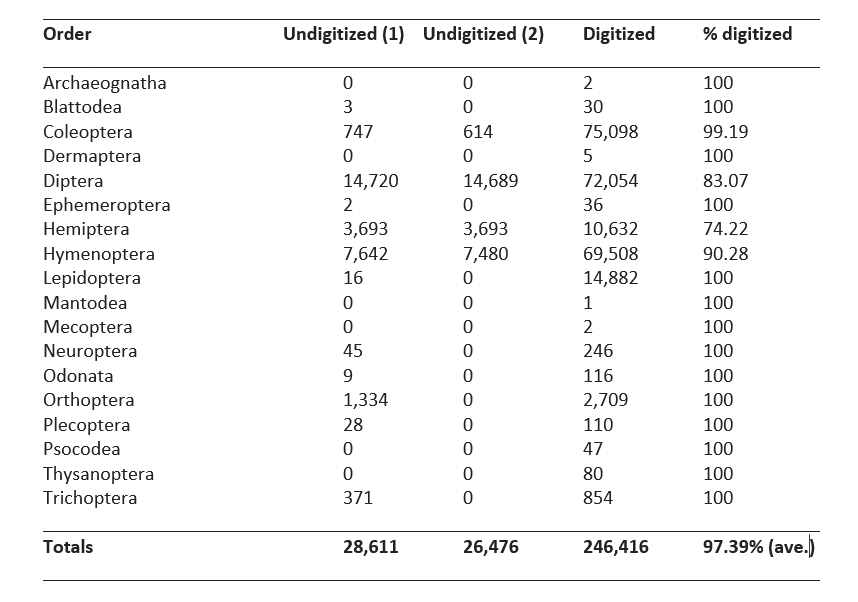
The results in Table 1 show that the UAM Insect pinned collection holds a minimum total of 272,892 specimens of which a little over 26,000 are not digitized (90.3% digitized). All small orders, those with fewer than 3,000 specimens, are 100% digitized. Of the larger orders, Lepidoptera is the most thoroughly digitized (100%) and Hemiptera the least (74.22%).
Table 1. List of pinned specimens in the UAM Insect Collection by order with initial (1) counts of undigitized specimens from Spring 2020, later current counts (2), current (8 Feb 2021) digital record totals, and percent digitization of each order.
Discussion
The most thoroughly digitized large order, Lepidoptera, at 100% was recently the focus of a large NSF-funded “Advancing Digitization of Biological Collections” grant titled LepNet (Seltmann et al. 2017), which was intended to complete digitization on all the UAM pinned Lepidoptera specimens. Coleoptera, the second most thoroughly digitized order at 99%, is the taxonomic focus of the curator (second author), and therefore has received more attention than others. However, Coleoptera (75,712 specimens) is only the third largest order in the collection behind Diptera (86,743 specimens) and Hymenoptera (76,988 specimens). The third most thoroughly digitized order, Hymenoptera, at 90% digitized, grew over the last 6 years from two separate unfunded donations. The first was a large collection of thousands of undigitized parasitoid wasps from the Dominique Collet collection; the second donation was of the Master’s thesis voucher specimens from University of Alaska Fairbanks graduate student Alexandria Wenninger. Together, these donations account for most of the undigitized Hymenoptera specimens. The fourth most thoroughly digitized order, Diptera, at ~83% digitized, has the largest total number of undigitized specimens (14,689), the bulk of which are from the USDA Palmer Agricultural Experiment Station and the Kathryn Sommerman biting fly collections of the mid 20th century. The least thoroughly digitized order, Hemiptera, at 74% digitized has many drawers of undigitized and unidentified Miridae and Cicadellidae (including many nymphs that are probably unidentifiable) from the USDA Palmer Agricultural Experiment Station collection. It is questionable whether these long series of old, unidentified and likely unidentifiable specimens are worth digitizing (or keeping for that matter). Their DNA might be of value, and probably the only way to identify most of them, but trying to obtain identifications in this manner would be very costly and a large percentage of specimens might fail to sequence due to degraded DNA (raising the cost per successful identification considerably).
The high rate of digitization in the UAM Insect Collection is due primarily to the use of the pro-active DBYL digitization protocol (Database Before You Label) as described in Sikes et al. (2017). This approach is typically an order of magnitude more efficient than retro-active digitization of already labeled and taxonomically sorted specimens. It ensures that specimens are ‘born digital’ with data captured at the collection event level, and have barcodes and database records before their labels are printed and before they are taxonomically sorted and identified. All the ~26,000 undigitized pinned specimens were prepared using the far less efficient method of typing data into a computer to generate labels (for those prepared in modern times), placing the labels on specimens, sorting taxonomically, and if any digitization happens at all, it is done by reading the data on the labels and typing those data back into a computer one specimen at a time. Assuming that growth of museum collections continues indefinitely into the future, then most specimens have yet to be collected. The last 200+ years of specimen collections will eventually become less than 10% of the material held in museums. If entomologists wish to maximize the value of limited research funding, they should adopt the DBYL protocol, or something similar, to ensure their specimens are ‘born digital’ and do not become a ten-fold cost burden on future generations.
Prior to this project we did not know the size of the pinned insect collection nor what percent was undigitized. Having completed this assessment, the UAM Insect Collection is now able to strategically target future digitization efforts to bring these specimens’ “dark data” into the light. We hope to conduct a similar assessment of the alcohol collection in the near future.
Acknowledgments
This work was completed in fulfillment of the University of Alaska Fairbanks Museum Research Apprenticeship (MRAP 288) course taken by the first author. We thank Kyle Campbell for encouraging the first author to take an MRAP course.
Author Contributions
The first author laid eyes on every pinned specimen while counting those that were undigitized, and drafted a report. The second author designed the project and helped with writing and analysis. The remaining authors worked on strategically digitizing specimens after the first pass had been completed and helped review the manuscript.
Literature Cited
Ceballos, G., Ehrlich, P.R., Barnosky, A.D., García, A., Pringle, R.M. and Palmer, T.M., (2015) Accelerated modern human–induced species losses: Entering the sixth mass extinction. Science Advances, 1(5), p.e1400253. https://doi.org/10.1126/sciadv.1400253
Heberling, J.M., Miller, J.T., Noesgaard, D., Weingart, S.B. and Schigel, D., (2021) Data integration enables global biodiversity synthesis. Proceedings of the National Academy of Sciences, 118(6). https://doi.org/10.1073/pnas.2018093118
Heidorn, P.B. (2008) Shedding light on the dark data in the long tail of science. Library Trends 57(2): 280–299. https://doi.org/10.1353/lib.0.0036
Nelson, G. and Ellis, S., 2019. The history and impact of digitization and digital data mobilization on biodiversity research. Philosophical Transactions of the Royal Society B, 374(1763), p.20170391. https://doi.org/10.1098/rstb.2017.0391
Raven, P.H., Miller, S.E. (2020) Here today, gone tomorrow. Science 370 (6513):149 https://doi.org/10.1126/science.abf1185
Seltmann, K.C., et al. (2017) LepNet: The Lepidoptera of North America Network. Zootaxa 4247 (1): 073–077 https://doi.org/10.11646/zootaxa.4247.1.10
Sikes, D.S. (2015) What is a specimen? What should we count and report when managing an entomology collection? Newsletter of the Alaska Entomological Society 8(1):3-8. http://www.akentsoc.org/doc/AKES_newsletter_2015_I.pdf
Sikes, D.S., Copas, K., Hirsch, T., Longino, J.T., Schigel, D. (2016) On natural history collections, digitized and not: a response to Ferro and Flick. ZooKeys 618: 145–158. https://doi.org/10.3897/zookeys.618.9986
Sikes, D.S. M. Bowser, K. Daly, T. T. Høye, S. Meierotto, L. Mullen, J. Slowik, J. Stockbridge. (2017) The value of museums in the production, sharing, and use of entomological data to document hyperdiversity of the changing North. Arctic Science 3: 498-514. http://dx.doi.org/10.1139/AS-2016-0038
Whitmore, V., Sikes, D.S., Haberski, A. (2020) University of Alaska Museum Insect Collection specimen count verification. Newsletter of the Alaska Entomological Society 13(1): 26-30 https://doi.org/10.7299/X7M9090P







