To facilitate the study of biodiversity, a number of software products are being collaboratively developed with researchers and projects. These websites, tools, and workflows take advantage of the data being digitized at US and global institutions and made available by iDigBio through our data services. Many other tools and services can be found through the Biodiversity Catalogue. If you have a great idea for using iDigBio data and web services, contact us or submit a proposal!
WordPress Leaflet Map Plugin Using iDigBio Data
iDigBio has collaborated with the Atlas of Ordovician Life project, part of the PALEONICHES-TCN to create a mapping plugin for WordPress that can generate maps of iDigBio specimen data on the fly. Leaflet Map enables map generation within WordPress webpages, and the WP Leaflet iDIgBio geojson data plugin developed by iDigBio enables an API query to iDigBio for specimen occurrence coordinates. An example of the plugin in use can be found here. More information about the plugin and the downloadable code can be found via GitHub at https://github.com/iDigBio/wp-leaflet-idigbio-geojson-data.
Working towards a Fitness-for-Use toolkit
The TDWG Biodiversity Data Quality Interest Group, a collaboration between data aggregators including iDigBio, researchers and collections data managers, is working towards developing standardized terminology and tools for assessing the research fitness-for-use for biodiversity data. Help with the development of data quality metrics by submitting your "user story" so that the team can better understand the specific needs of the community of biodiversity data users.
riDigBio: an R interface to the iDigBio Data API
Familiar with R? The riDigBio package is a great way to interface with the data shared by iDigBio. Contributions to this R package can be made via GitHub.
FreshData and Effechecka
A collaboration between iDigBio, Encyclopedia of Life (EOL), and Jorrit Poelen (freelance software engineer, GloBI) is resulting in two tools using iDigBio primary biodiversity data and other data sources: Effechecka is a taxonomic checklist generator which uses spatial and trait data, and FreshData enables researchers to receive notification as to when new biodiversity data, based on a particular query string, has become available from various data sources, such as iDigBio. Code for both these applications are available via GitHub. iDigBio is supporting these collaborative projects by providing hardware, bandwidth, and data archives, EOL provided funding and use cases, and Jorrit Poelen developed the software and data architecture to build the prototypes. For more information about the GUODA (Global Unified Open Data Access) project behind FreshData and Effechecka, see this poster!
SuAVE: Survey Analysis via Visual Exploration
 iDigBio are working with Ilya Zaslavaky, Director of the Spatial Information Systems Lab at SDSC, to develop a SuAVE extension for curating and exploring large collections datasets, support domain standards, interface with taxonomies, and make it available through an analysis platform. SuAVE uses technical approaches from image analytics, faceted search, and online map navigation as an online tool for exploratory data analysis.
iDigBio are working with Ilya Zaslavaky, Director of the Spatial Information Systems Lab at SDSC, to develop a SuAVE extension for curating and exploring large collections datasets, support domain standards, interface with taxonomies, and make it available through an analysis platform. SuAVE uses technical approaches from image analytics, faceted search, and online map navigation as an online tool for exploratory data analysis.
 Publishing biodiversity data made easy (and trackable) with Pensoft!
Publishing biodiversity data made easy (and trackable) with Pensoft!
The Biodiversity Data Journal (BDJ) now allows streamlined and direct import of biodiversity occurrence records from iDigBio using the ARPHA Publishing Platform. Pensoft and BDJ are looking for users, use cases, and collaborators for their new (revolutionary!) writing tool. Take a look at the blog post and webinar from Pensoft for more information, and ontact them at info@pentsoft.net for more information and to get started!
ePANDDA
 Enhancing Paleontological and Neontological Data Discovery (ePANDDA) is a new API being developed for geoscience database connectivity. There are already numerous geological, paleontological, and biological databases available for an array or geoscience research. ePANDDA will connect three established and well-supported database initiatives:
Enhancing Paleontological and Neontological Data Discovery (ePANDDA) is a new API being developed for geoscience database connectivity. There are already numerous geological, paleontological, and biological databases available for an array or geoscience research. ePANDDA will connect three established and well-supported database initiatives:
- The Paleobiology Database (PBDB, paleontological, literature based)
- iDigPaleo (paleontological, specimen-based)
- iDigBio (neontological, specimen-based).
This project is creating a seamless search and discovery of data among the three. The open architecture of ePANDDA will encourage the inclusion of other types of online datasets, for example core samples, isotopic analyses, or paleomagnetic samples.
BiotaPhy
The NSF-funded project "Connecting resources to enable large-scale biodiversity analyses", a.ka. BiotaPhy is developing a cyberinfrastructure framework linking diverse data (phylogeny, morphology, ecology, fossils, geography, and climate) across several data-driven community research tools. These community tools include Open Tree of Life, which will serve as the framework to which all other biological data – traits, genes, genomes, and especially specimens – will be linked, iDigBio (ultimately ~1 billion museum specimens that carry locality data and their implicit ecological information), Lifemapper (geospatial data, modeling, and analyses), and Arbor (diverse analytical tools leveraging myriad input data).
PhyloJIVE (Phylogeny Javascript Information Visualiser and Explorer)
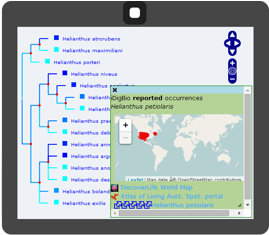 Solutions to fundamental questions about biodiversity require a new approach that integrates across phylogeny, biogeography, geology, and paleobiology. PhyloJIVE, developed by Garry Jolley-Rogers, Joe Miller, and Temi Varghese, integrates biodiversity data with phylogeny. Through PhyloJIVE, occurrence records can be viewed in a phylogenetic context, and user-supplied character data can be visualized on the phylogeny. Exploration of the linkages between phylogeny, distributions, and character states can lead to new understanding of biodiversity. PhyloJIVE is currently being upgraded to PhyloLink by iDigBio collaborators, Atlas of Living Australia.
Solutions to fundamental questions about biodiversity require a new approach that integrates across phylogeny, biogeography, geology, and paleobiology. PhyloJIVE, developed by Garry Jolley-Rogers, Joe Miller, and Temi Varghese, integrates biodiversity data with phylogeny. Through PhyloJIVE, occurrence records can be viewed in a phylogenetic context, and user-supplied character data can be visualized on the phylogeny. Exploration of the linkages between phylogeny, distributions, and character states can lead to new understanding of biodiversity. PhyloJIVE is currently being upgraded to PhyloLink by iDigBio collaborators, Atlas of Living Australia.
Lifemapper
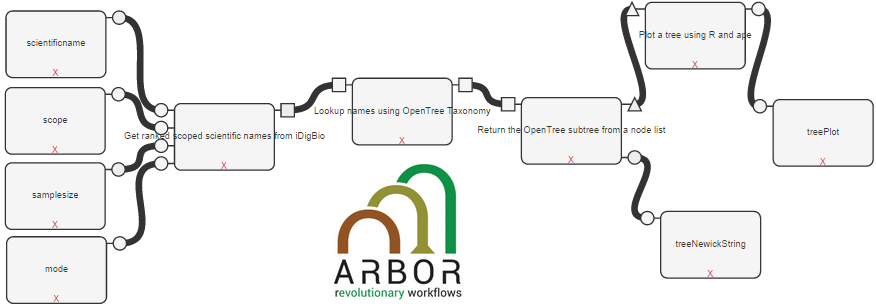
Arbor Workflows
Arbor brings comparative biology into the age of big data. This software is an extendable platform for analyzing and visualizing evolutionary processes across the tree of life. Arbor provides an integrated modeling framework that will enable researchers to answer evolutionary questions using very large phylogenetic trees and diverse types of data. The goal of Arbor workflows is to provide biologists with an immediate and intuitive interface for testing evolutionary questions across the tree of life and to enable them to communicate their findings to scientific and general audiences. A series of tutorials about how to use the tools can be found here.
Biospex
 BIOSPEX is a basecamp for launching, advertising, and managing targeted efforts to digitize vouchered biological specimens. Motivations to digitize collections data can vary widely, from the museum curator to the descendants of a collector reconstructing their ancestor's steps to an environmental group interested in the health of a local water body. BIOSPEX enables each of these interested researchers to package their projects in one or a series of digitization expeditions, launch the expeditions as crowdsourcing tools, and widely recruit others to participate. In the end, the new data can be downloaded AND returned to the museum that curates the physical specimen.
BIOSPEX is a basecamp for launching, advertising, and managing targeted efforts to digitize vouchered biological specimens. Motivations to digitize collections data can vary widely, from the museum curator to the descendants of a collector reconstructing their ancestor's steps to an environmental group interested in the health of a local water body. BIOSPEX enables each of these interested researchers to package their projects in one or a series of digitization expeditions, launch the expeditions as crowdsourcing tools, and widely recruit others to participate. In the end, the new data can be downloaded AND returned to the museum that curates the physical specimen.