Moving entire pinned insect collections into the digital realm in one fell swoop
 Contributed by: Mark Hereld
Contributed by: Mark Hereld
Senior Experimental Systems Engineer
Mathematics and Computer Science
Argonne National Laboratory
http://www.mcs.anl.gov/person/mark-hereld
I am working with a small team at Argonne National Laboratory to develop methods for very high-speed digitization of large collections of pinned insects. These collections represent a significant societal investment in research and applied environmental science. We are not alone in thinking that if these assets were available in digital form, new and amazing science would be enabled while also creating novel opportunities for outreach and education.
Some researchers have chosen to focus on targeted subsets of a collection in order to address a specific scientific question. We take a different approach. Our interest is in digitizing the entire collection to create a homogeneous dataset that can be used at once in a wide variety of scientific inquiries. Unfortunately, available methods for specimen digitization would require decades and more to realize this goal.
Taking the collection at the Field Museum of Natural History (FMNH) as a use case, a pipeline to digitize the entire collection of 4.5 million specimens in around 1 to 2 years imposes a limit of only a few seconds on average to capture each specimen! In our research and development effort, we are hoping to find new and efficient approaches to this challenge by virtue of our experience in instrument design, computer vision and image analysis, and the techniques employed in parallel computing for optimizing computational pipelines.
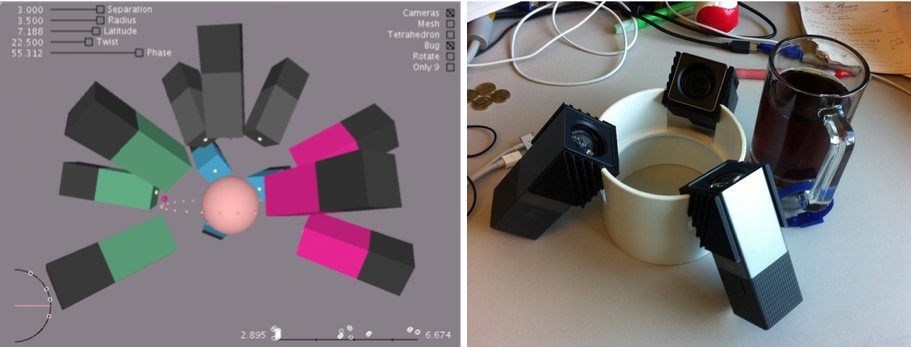
Figure 1. Our first design of a modular multi-camera 3D capture rig is built out of four modules, each outfitted with three cameras. The sketch on the left shows how four color-coded modules are arranged to capture many views of the specimen at once. The image on the right shows one of those modules as built.
In our approach, we try to minimize the time it takes to handle and arrange the specimen in preparation for recording the information on its labels. We have estimated that the overwhelming majority of specimens in the FMNH collection (perhaps 98%) are small enough that every part of each label on the pin will be visible from some angle. We reason that by taking pictures of the specimen from many angles, it should be possible to completely avoid handling the specimen except when removing it from and returning it to its drawer. And, by taking all of those images at once with many cameras arrayed around the specimen, instead of using one camera to sequentially capture images from different positions, we can effectively digitize the specimen by taking one multi-camera snapshot! These data can then be analyzed to identify and combine fragments of printed label text to recreate whole labels.
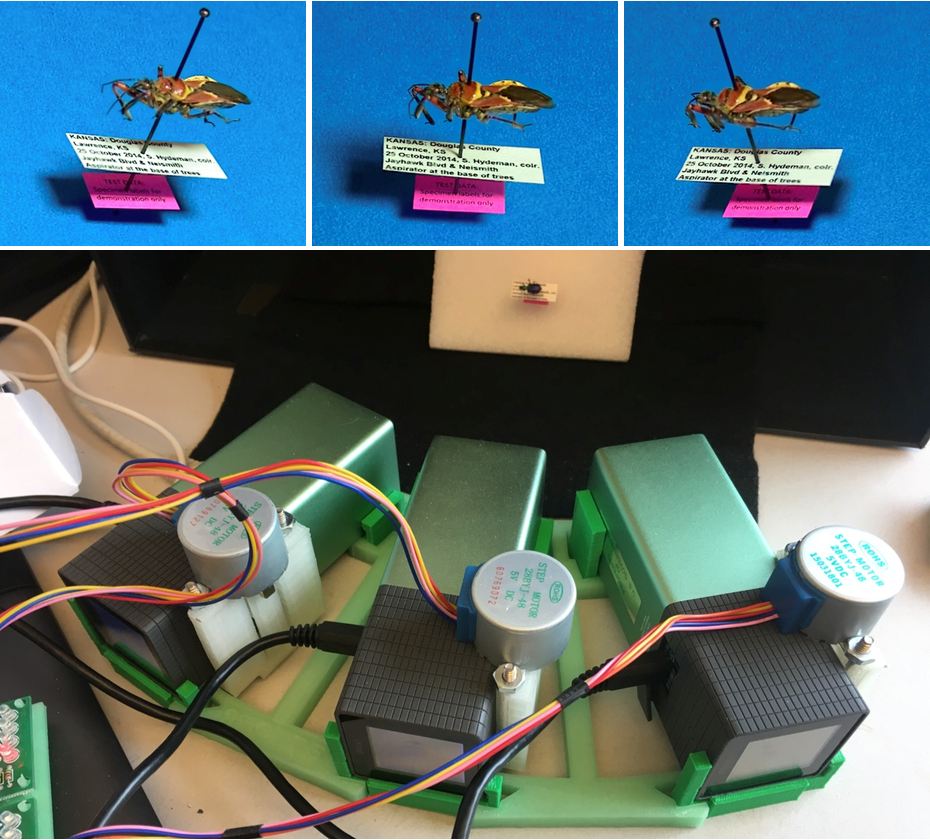
Figure 2. An example of a multi-camera snapshot produced by a simple 3-camera system (pictured) with side-by-side cameras arrayed tightly around a central point.
And so, we have worked through a number of multi-camera designs aimed at solving this problem efficiently and economically. Figure 1 shows one such design which arrays twelve cameras around the specimen. Figure 2 shows a smaller system that we designed for digitizing labeled sample vials (instead of pinned insects), and have used to quickly capture test data to help us develop the algorithms for converting fragmentary images of the labels into face on views ready for transcription by, for example, optical character recognition (OCR) software.
Recreating virtual whole labels from the multi-view data is an interesting and challenging problem. Part of the process is illustrated in Fig. 3 for several examples. First the labels are automatically identified in the image. Edges and corners on each label are discovered and combined with the visual information in the text to define a coordinate grid. With that grid, the label area can be mathematically deformed to undo the effects of perspective, resulting in a face on view (not shown) of the visible portion of the label. Note that in each of these images, portions of the labels are occluded by the insect or another label. The effects of these missing or contaminated portions of each view are removed when the fragments from multiple views are combined.

Figure 3. Illustration showing the result of automatically recognizing features in the label to arrive at a coordinate grid on its face.
The systems described to this point have helped us to develop our ideas, but we have found the cameras in them to be impractical. Consequently, our most recent design (Fig. 4) uses very inexpensive and widely available camera and controller components that are more readily integrated into our high-speed data acquisition system. We are beginning to test this design with half of the planned twelve cameras in place. The software that captures images from all of the cameras and archives the results to a database allows us to take a multi-camera snapshot every second. The full complement of cameras will be live shortly and ready for a speed test in the coming weeks. Our plan for this test is to demonstrate sustained high-throughput capture of the imagery needed to digitize the contents of the labels. Exciting times, indeed!

Figure 4. Our most recent design, in the top panel, can accommodate many cameras (36 as shown, 48 for full circle). The “abstract sculpture” in the center of the diagram is a schematic representation of the sample volume, approximately 2” x 2” x 2”. The lower left panel shows the twelve-camera configuration of the 3D printed rig before the cameras were installed, including a clear avenue for the conveyor belt. The lower right panel shows one of the very compact cameras (1” x 1”) on its 3D printed custom mounting bracket.
Learn More
- Mark Hereld, Nicola J. Ferrier, Nitin Agarwal and Petra Sierwald, "Designing a High-Throughput Pipeline for Digitizing Pinned Insects," 2017 IEEE 13th International Conference on e-Science (e-Science), BigDig 2017 Workshop, Auckland, 2017, pp. 542-550. doi: 10.1109/eScience.2017.88 (pdf) (slides)
- Perspectives for combining 3D and mass-digitisation - Mark Hereld (Mathematics and Computer Science Division, Argonne National Laboratory, Argonne, Illinois) (video) (pdf)
- BigDig: High Throughput Digitization for Natural History Collections, a workshop held in Auckland, NZ, October 2017.







