Data Ingestion Guidance: Difference between revisions
| Line 153: | Line 153: | ||
*'''continent''': For details see: http://rs.tdwg.org/dwc/terms/#continent | *'''continent''': For details see: http://rs.tdwg.org/dwc/terms/#continent | ||
*'''dynamicProperties''': when including data in the dynamicProperties field, please use JSON format. For details see: http://rs.tdwg.org/dwc/terms/#dynamicProperties. | *'''dynamicProperties''': when including data in the dynamicProperties field, please use JSON format. For details see: http://rs.tdwg.org/dwc/terms/#dynamicProperties. | ||
Other fields for completeness include, and can be configured as defaults that can be set up in IPT: | Other fields for completeness include, and can be configured as defaults that can be set up in IPT for all records: | ||
*'''basisOfRecord'''="PreservedSpecimen" or "FossilSpecimen". For details see: http://rs.tdwg.org/dwc/terms/#basisOfRecord | *'''basisOfRecord'''="PreservedSpecimen" or "FossilSpecimen". For details see: http://rs.tdwg.org/dwc/terms/#basisOfRecord | ||
*'''type'''="PhysicalObject" For details see: http://rs.tdwg.org/dwc/terms/#type | *'''type'''="PhysicalObject" For details see: http://rs.tdwg.org/dwc/terms/#type | ||
Revision as of 14:47, 13 January 2015
Data Ingestion Workflow
Working copy 1.1 (June 2014)
Audience: iDigBio data ingestion staff
This is the process description for
- the iDigBio staff to follow to assure that data are successfully and efficiently moved from data provider to the portal, available for searching.
- data providers to follow to assure that data are efficiently and accurately provided to the iDigBio staff.
First step to becoming a data provider
Sending your data to us is as simple as sending us an email to data@idigbio.org to say where to pick it for ingestion. If you need help compiling it into the acceptable formats, then get in touch with us to express your interest, and we'll help with what you.
Contact info
If you need assistance related to data ingestion, contact data@idigbio.org.
Register your data
iDigBio accepts specimen data and related media from any US-based institution. If you are ready to discuss providing data to iDigBio, contact data@idigbio.org to register your interest and begin the process of preparing your data for ingestion. If you have a Darwin Core Archive (DwC-A), getting your data ingested with iDigBio could be as easy as telling us the RSS feed address on your network. Information about setting up an RSS feed can be found here: Setting up an RSS feed
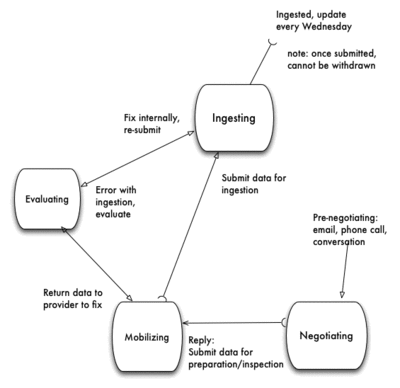
Process terminology
Processing steps, each step has a start and an end, signifying that it has moved to the next step.
- negotiating - in the process of determining provider's interest in data ingestion
- begins with email invitation to providers (in institutions, aggregators) to invite them to send their data to iDigbio specimen data portal
- open a Redmine ticket in project=Data Mobilizing
- ends with data exported by provider, ready for inspection and ingestion.
- mobilizing - in the process of evaluating data being fit for ingestion
- begins with provider exported data and cursory inspection
- fill in this table with provider info: eml.xml, unless there is a good eml.xml file available (e.g., from a DwC Archive)
- ends with data passing inspection and passing to ingesting state, Redmine ticket changes to assignee=cyberinfrastructure team
- ingesting - in the process of ingesting provider's data
- begins with Redmine ticket change to assignee=cyberinfrastructure team
- ends with
- data successfully ingested, ready for consumption in the portal
- report sent back to data mobilizing staff
- report sent to provider. Reference: Publishers Report
- Redmine ticket set to Status= Closed
- evaluating - in the process of evaluating a failure to be ingested
- begins with ingestion failure
- evaluate ingestion failure, if data error - send it back to mobilizing state for corrections or
- evaluate ingestion failure, if ingestion error - make corrections
- ends with data re-submission to ingesting state
- begins with ingestion failure
Data requirements for data providers
Below are what we ask of the data to make it easily searchable in the cyberinfrastructure we provide.
There are 3 kinds of data files to submit for ingestion:
- specimen data with dataset metadata
- media data related to and attached by reference to specimen records with metadata (use of dwc:associatedMedia in the occurrent/specimen data file is not viewed as sending media)
- media files - e.g., non-archival .jpgs (see acceptable format here: https://www.idigbio.org/content/idigbio-image-file-format-requirements-and-recommendations-1)
Packaging for specimen data
In order of preference:
- DwC-A (Darwin Core Archive) produced by IPT on a RSS feed. IPT is available at: https://code.google.com/p/gbif-providertoolkit/
- Custom DwC-A on an RSS feed produced by Symbiota
- Custom CSV or TXT (save the data in UTF-8 format to preserve diacritics in people and place names), this option for sending only specimen data or only media data (DwC-A packaging required when sending both specimen and media data)
- Custom RSS feed following the guidance at: iDigBio RSS specification
- Standard DwC-A uses field names from:
- Darwin Core: http://rs.tdwg.org/dwc/terms/
- Audubon Core: http://terms.tdwg.org/wiki/Audubon_Core_Term_List
- A custom CSV allows providers to send data beyond standards such as Dublin Core and Darwin Core. For example, providers can send tribe taxonomic information in the field "idigbio:tribe". While creating additional fields, use field names that follow DwC format (camel case), additionally, consult the MISC field names (local iDigbio extensions to DwC). The host association terms are an example of an extension found in the MISC. Use the XML style field names that include the domain of the schema, e.g., dwc:termName, ac:termName. Non-standard field names are indexed and available through search API.
Special note to data aggregators
Note to aggregated data providers (e.g., California Consortium of Herbaria (CCH), Calbug, Tri-Trophic TCN (TTD), Consortium of Pacific Northwest Herbaria (CPNW)):
When providing us access to your data, we highly encourage you to provide your aggregated data one provider at a time, each in their own Darwin Core archive, with their own list of contacts in their separate EML file. iDigBio is moving towards providing data quality feedback, data correction, annotations, and other value-added information back to the providers and thus we want individual contact info for each source provider where possible. The hope is that the information could be re-integrated at the source so that higher quality data would be in place at the provider as well as be available to downstream data consumers such as iDigBio and GBIF.
However if that is not possible or desirable, we still welcome your aggregated data as one monolith.
With the interest of people/researchers using your data in the aggregate, e.g., EOL, we encourage you to homogenize the rights information you provide. We recommend CC0 (http://creativecommons.org/publicdomain/zero/1.0/).
Sending data to iDigBio
- An RSS feed to a DwC-A for ready access and update is our preference
- Email the files to us
Specimen metadata
- Each specimen record should have a unique (within the dataset) identifier in the dwc:occurrenceID field. When the ingestion software detects duplicate identifiers, the duplicated records are flagged as an error and are not ingested. This is the number one reason for records to be rejected. Identifiers, if not GUIDSs or specifically UUIDs, are what is typically called the DwC (Darwin Core) triplet:
<dwc:institutionCode>:<dwc:collectionCode>:<dwc:catalogNumber>
example with a prefix (lowercase is preferred in the prefix):
urn:catalog:TNHC:Herpetology:122
Further examples include:
- a simple / bare UUID:
f47ac10b-58cc-4372-a567-0e02b2c3d479
- a UUID using URI syntax: (lowercase is preferred in the prefix)
urn:uuid:f47ac10b-58cc-4372-a567-0e02b2c3d479
- an Archival Resource Key (ARK):
ark:/87286/f47ac10b-58cc-4372-a567-0e02b2c3d479
- any of the various GUID types defined in TDWG Standard 150 (http://www.tdwg.org/standards/150/).
Complete attribution and licensing
In order for each provider's data to be correctly attributed when found on the iDigBio portal, the following are important to complete:
- Fill in your official institution code (dwc:institutionCode) and collection code (dwc:collectionCode)
- check your entry in grbio.org and make sure it is current and complete : http://grbio.org/
- here: Repositories: http://grbio.org/find-biorepositories
- here: Institutional collections: http://grbio.org/find-institutional-collections
- make sure you have used the same institutionCode and collectionCode in GRBio, and your EML/IPT dialog
- check your entry in grbio.org and make sure it is current and complete : http://grbio.org/
- Go to http://GRBio.org to get their Cool URI value for your institution in the alternateIdentifier field in the EML dialog ( (e.g., http://biocol.org/urn:lsid:biocol.org:col:15587)). Store in the dwc:institutionID and dwc:collectionID fields
- Fill in the DwC global-to-the-dataset DwC record-level fields for intellectual property and licensing, e.g., dcterms:rights, dcterms:rightsHolder and dcterms:accessRights or use the global EML-based dwc:intellectualRights field.
- Use the field dcterms:bibliographicCitation, e.g., Ctenomys sociabilis (MVZ 165861) for the correct attribution string for each record.
- Check our intellectual property policy for complete details.
dcterms:rights
- any actual rights statements (IP, or otherwise), and any licenses associated with the data sets (e.g., CC0), chosen from the Creative Commons options. Any right or license will appear with each record it covers.
dcterms:rightsHolder
- will be blank unless the publisher has content in this field that they have entered on their own. If the publisher chooses to put their institution name, or an individual name in this field is up to them. This tends to be a blank field.
dcterms:accessRights
- is where the terms of use should be placed, things such as you have to attribute us or provide us with a final copy of a given product. It will be blank unless the provider has entered content at the source and on their own.
dwc:intellectualRights example
- institution-name data records may be used by individual researchers or research groups, but they may not be repackaged, resold, or redistributed in any form without the express written consent of a curatorial staff member of the institution-name. If any of these records are used in an analysis or report, the provenance of the original data must be acknowledged and the institution-name notified. The institution-name and its staff are not responsible for damages, injury or loss due to the use of these data.
Several examples of the use of public domain, recommended for specimen data:
dcterms:rights = http://creativecommons.org/publicdomain/mark/1.0/ xmpRights:webStatement = http://creativecommons.org/publicdomain/mark/1.0/ dc:rights = Public Domain xmpRights:owner = Public Domain
Some further guidance on this subject: '...when you are completing the metadata in the IPT, under Additional Metadata, it is important to consider the licensing and rights that you may wish to publish the data under. There are a couple of interesting articles describing the reasoning behind the Creative Commons licenses, http://creativecommons.org/licenses/, at the following URLs:
- http://www.canadensys.net/2012/why-we-should-publish-our-data-under-cc0
- http://peterdesmet.com/posts/illegal-bullfrogs.html
- http://peterdesmet.com/posts/analyzing-gbif-data-licenses.html
It may also be useful to read the Creative Commons Wiki on using Creative Commons licenses on data. http://wiki.creativecommons.org/Data" (ref D. Bloom)
Permission to ingest
- the provider needs to have permission to submit their data
Data recommendations for optimal searchability
- Dates: put dates in ISO 8601 format, i.e., YYYY-MM-DD, e.g., 2014-06-22. The critical element in this date is a four character year. e.g., http://rs.tdwg.org/dwc/terms/#eventDate.
- Meters: put elevation in METERS units in the elevation field without the units (e.g., the fields dwc:minimumElevationInMeters and dwc:maximumElevationInMeters already assume the numeric values are in meters, so there no need to include the units with the data).
- Escapes: do not use unescaped newline characters.
- No '0': do not export '0' in fields to represent no value, e.g., lat or lon. This caution applies to '?', 'NA', '00/00/0000' and any other placeholder value.
- decimalLatitude & decimalLatitude: make sure lat and lon coordinates are in decimal, and no N, S, E, W. For details see: http://rs.tdwg.org/dwc/terms/#decimalLatitude.
- genus, specificEpithet, infraspecificEpithet & taxonRank: parse taxon ranks.
- scientificName: combine taxon ranks into the identification value.
- vernacularName: include common names for broader audience fundability. For details see: http://rs.tdwg.org/dwc/terms/#vernacularName
- higherClassification: include parsed higher taxonomy classification, at least kingdom and family, and the intervening ranks if possible. For details see: http://rs.tdwg.org/dwc/terms/#higherClassification.
- countryCode: include a 3 character countryCode from here: http://en.wikipedia.org/wiki/ISO_3166-1_alpha-3. For details see: http://rs.tdwg.org/dwc/terms/#countryCode.
- continent: For details see: http://rs.tdwg.org/dwc/terms/#continent
- dynamicProperties: when including data in the dynamicProperties field, please use JSON format. For details see: http://rs.tdwg.org/dwc/terms/#dynamicProperties.
Other fields for completeness include, and can be configured as defaults that can be set up in IPT for all records:
- basisOfRecord="PreservedSpecimen" or "FossilSpecimen". For details see: http://rs.tdwg.org/dwc/terms/#basisOfRecord
- type="PhysicalObject" For details see: http://rs.tdwg.org/dwc/terms/#type
- language= "en" For details see: http://rs.tdwg.org/dwc/terms/#language
Packaging for images / media objects
- Firstly, adding a field in the occurrence file for associatedMedia is not the way to include media with a specimen record.
- Each media record should have a unique (within the dataset) identifier in the dcterms:identifier field.
- If submitting media records with specimen data records, here are critical fields to fill in:
- If you have a UUID GUID for your image records, then assign it to the ac:providerManagedID field.
- sample
- id = identifier of the specimen record
urn:catalog:institutionCode:collectionCode:catalogNumber
- identifier = id of the media record - needs to be unique within Audubon Core file
urn:catalog:institutionCode:collectionCode:Image:catalogNumber
- accessURI = link to the media file
http://bgbasesrvr.univ.edu/DATABASEIMAGES/LONN00000001.JPG
- providerManagedID =
urn:uuid:32e5da5d-c747-435c-a368-07d989259bf4 (optional)
- id = identifier of the specimen record
- If you are not using IPT, generate a meta.xml file by hand and package up the files in a DwC-A like format. (No eml.xml required).
- sample
- If you have a UUID GUID for your image records, then assign it to the ac:providerManagedID field.
- Use Audubon Core terms, http://terms.tdwg.org/wiki/Audubon_Core_Term_List, with one record to go with each media record. The more you can flesh out the details of the image, the more likely it will be to be highly retrievable. The best practice is to use the taxonomic and geographic fields to capture as much information as possible when only media are given to iDigBio.
- Just like permission of catalog records, the media records need to be provided freely and with permission, and each record should have at least Creative Commons permission = "CC BY" unless it is in the public domain.
- a sample of an Audubon Core file
- The media records represent a one-to-one relationship between the media object (the fit-for-display best quality JPG, in the case of images, for example) and the specimen record. There is no need to include links to any other forms of the media, for example an enclosing webpage.. Below is some guidance on handling special cases. If none of these media attachment rules make sense to you, please get in touch with us for further assistance.
Best practice for getting Audubon Core images linked to specimen records - special cases
| Relationship | Supported by | Core Type | Extensions |
|---|---|---|---|
| One-specimen-record-to-many-media files | IPT 2.1/Custom DwC-A | Specimen (DwC) | Audubon Core |
| Many-specimen-records-to-one-media file | IPT 2.2/Custom DwC-A | Audubon Core | Specimen (DwC) |
| Many-specimen-records-to-many-media files | IPT 2.1/Custom DwC-A | Specimen (DwC) | Audubon Core + Relationship |
Keep in mind that:
- DwC-A is a set of files: a core type + a number of extensions
- All files/tables (core or extension) need to have a unique identifier
Notes on getting data from EMu into a Darwin Core Archive
The cookbook recipe is provided by Larry Gall, Yale Peabody Museum
- Schedule a regular cron job to export out of EMu each night the Darwin Core fields that are part of a shadow schema built into EMu (this could be part of part of the nightly backup/dump processing).
- Later in the cron that exported file is the input to a script that does some additional field cleanup and parsing, and writes a delimited file which in turn is scped to your IPT server
- Use this copied, delimited file to reinstantiate MySQL tables that are the source for the IPT (via truncate table x and load data local infile x).
Error handling
When data are received from the provider during the mobilizing process step, they are evaluated for fitness. Once the evaluation is successful, the ingestion process moves from mobilizing to ingesting, and the data are submitted to the ingestion scripts by the cyberinfrastructure staff. If an error condition occurs, the staff evaluate whether it is a script error or a data error. If it is the latter, the staff sends an email to the mobilizing staff who may contact the provider for changes. When the errors have been addressed, the mobilizing staff re-submit the data to the ingesting staff.
Sample scenarios of data transformations to prepare data for ingestion
- Example preparing specimen data from Illinois Natural History survey (INHS) fish collection from FileMakerPro
- Example of transformations on InvertNet image metadata dataset
Advertising your data on iDigBio on your website
We encourage you to post a link on your institution's website informing users that they will also find your data on iDigBio's portal.
Please look here for logo material: https://www.idigbio.org/wiki/index.php/IDigBio_Logo
and consider making the link to be to your publishers page, something like:
https://www.idigbio.org/portal/recordset/c50755ff-ca6d-4903-8e39-8b0e236c324f
where the UUID on the end of this link belongs to your recordset. The link to your recordset can be found here: iDigBio publishers
Additional references
If you want to learn about acceptable Creative Commons licenses in iDigBio:
Data ingestion report, progress so far
- https://www.idigbio.org/wiki/index.php/Data_Ingestion_Report
- https://www.idigbio.org/portal/publishers
Provider assistance
- How to use the image ingestion appliance and link to specimen records : image ingestion cheatsheet
- How to generate a UUID GUID in an Excel spreadsheet
- How to use IPT to include images in your Darwin Core Archive (using Audubon Core)
- Setting up an RSS feed
- If you are interested in the terms used by providers in their data so far, including non-DwC ones, this is a snapshot as of 05/20/2014: