IDigBio API v1 Examples: Difference between revisions
No edit summary |
|||
| (73 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
[[Category:Documentation]] | [[Category:Documentation]] | ||
This document is a list of examples for using the | {{Caution|The v1 API has been superseded by [https://github.com/idigbio/idigbio-search-api/wiki iDigBio API v2 / iDigBio Search API]. API users should migrate to using the most recent version of the API. This page remains for historical reference.}} | ||
This document is a list of examples for using the [[IDigBio API]] but may not be comprehensive to cover every feature of the API. See [[iDigBio API v1 Specification]] for full documentation on the API including all of the available endpoints and parameters. | |||
=== Viewing JSON Output === | === Viewing JSON Output === | ||
Most of the examples provided here will be shown using the <code>curl</code> command-line tool. | Most of the examples provided here will be shown using the <code>curl</code> command-line tool. The dollar sign <code>$</code> is an indication of a command prompt where something is typed into a shell or terminal. To run the commands, the <code>$</code> should be omitted. | ||
Here are some of the more commonly used curl options in our examples (for descriptions and additional usage information, see the curl man page): | |||
<pre> | <pre> | ||
| Line 13: | Line 17: | ||
... | ... | ||
-s, --silent | -s, --silent | ||
-v, --verbose | -v, --verbose | ||
-d, --data <data> | -d, --data <data> | ||
... | ... | ||
</pre> | </pre> | ||
Many of the examples will also include "pipes" to chain multiple commands together. The [http://en.wikipedia.org/wiki/Pipeline_(Unix) Wikipedia article on Unix software pipelines] may be a useful reference. | |||
==== Javascript Object Notation (JSON) Quick Intro ==== | |||
Javascript Object Notation ([http://en.wikipedia.org/wiki/JSON JSON]), is an open standard format that uses human-readable text to transmit data objects consisting of attribute–value pairs. JSON has become a common format for interchanging data via APIs and especially modern Web Services. Effective use of the iDigBio API will require some understanding of JSON. | |||
A simple JSON document with two fields and two data values would look something like this: | |||
<pre> | <pre> | ||
{ | |||
"field1" : "value_a", | |||
"field2" : "value_b" | |||
} | |||
</pre> | |||
{" | If we were storing data about John Smith, age 50, we might represent it as the following JSON document: | ||
<pre> | |||
{ | |||
"lastName" : "Smith", | |||
"firstName" : "John", | |||
"age" : "50" | |||
} | |||
</pre> | </pre> | ||
It may be useful to | A short JSON document is returned by the iDigBio API when calling the HTTP "GET" method (curl's default) against the base URL (http://api.idigbio.org): | ||
<pre> | |||
{ | |||
"v1" : "http://api.idigbio.org/v1/", | |||
"check" : "http://api.idigbio.org/check", | |||
"v0" : "http://api.idigbio.org/v0/" | |||
} | |||
</pre> | |||
It may be useful to inspect the API's JSON document outputs using a "pretty printing" feature of some kind. This will make it more readable by humans and also allow it to be more easily filtered using tools such as <code>grep</code>. Two common tools for this available on Unix/Linux are python's json.tool module or the json_pp command-line Perl script (JSON::PP). In typical Unix fashion, these tools can receive input from a pipe and be used to chain multiple commands together. | |||
There are also ways of viewing JSON inside of a web browser. | |||
Most of our examples in this document will include piping the JSON document into json_pp. | |||
==== JSONView Web Browser Extension ==== | |||
For viewing of JSON results in a browser, one may wish to use the [http://jsonview.com/ JSONView Extension] which is available for Firefox and Chrome. | |||
Here is a screenshot of the output using the JSONView extension after requesting the API base URL: | |||
[[File:JSONView_browser_extension_example.png|border|JSONView extension allows easier viewing in a web browser]] | |||
==== json.tool (Python) ==== | |||
Current installations of Python typically include a json module in the standard installed libraries. | |||
<pre> | <pre> | ||
$ curl -s http://api.idigbio.org/ | python -m json.tool | $ curl -s http://api.idigbio.org/ | python -m json.tool | ||
| Line 54: | Line 85: | ||
</pre> | </pre> | ||
==== json_pp ==== | ==== json_pp (Perl) ==== | ||
Similarly, modern Perl installations typically include the json_pp script or it can be aquired from [http://search.cpan.org/perldoc/json_pp CPAN]. | |||
<pre> | <pre> | ||
$ curl -s http://api.idigbio.org/ | json_pp | $ curl -s http://api.idigbio.org/ | json_pp | ||
| Line 64: | Line 98: | ||
</pre> | </pre> | ||
==== simplejson.tool (Python) ==== | |||
If a Python installation is misssing the json standard library, the separate simplejson package may solve the problem. | |||
The simplejson.tool module can be invoked in the same way as json.tool: | |||
<pre> | |||
$ curl -s http://api.idigbio.org/ | python -m simplejson.tool | |||
{ | |||
"check": "http://api.idigbio.org/check", | |||
"v0": "http://api.idigbio.org/v0/", | |||
"v1": "http://api.idigbio.org/v1/" | |||
} | |||
</pre> | |||
Older versions of python also may require a more complicated syntax to invoke the module from the command line. | |||
<pre> | <pre> | ||
| Line 83: | Line 124: | ||
</pre> | </pre> | ||
=== | === Three ways to fetch a specific record based on iDigBio UUID === | ||
The following three examples intend to illustrate the various methods for looking up a unique record in iDigBio | The following three examples intend to illustrate the various methods for looking up a unique record in iDigBio. Each record that is added to iDigBio is assigned a Globally Unique Identifier (GUID) of the UUID type. | ||
The data returned by the Elasticsearch interface may differ from the raw API record, especially with regards to field names, since the Elasticsearch data has been normalized and indexed for consumption by the search portal. | The data returned by the Elasticsearch interface may differ from the raw API record, especially with regards to field names, since the Elasticsearch data has been normalized and indexed for consumption by the search portal. | ||
For these three examples, consider the | Regardless of whether the record's data is retrieved by using the API or by using Elasticsearch, the piece of information used to identify a record is the iDigBio UUID. | ||
For these three examples, consider the specimen record identified by the following iDigBio UUID: | |||
<pre> | <pre> | ||
| Line 97: | Line 140: | ||
==== Use the API ==== | ==== Use the API ==== | ||
A complete record that was provided to iDigBio is stored in the API and can be retrieved by referencing the iDigBio UUID ("idigbio:uuid") at the appropriate endpoint. | |||
The following | Let us say that we have already located the specimen record for a particular Curculionidae specimen (a family of weevils). The specimen record for our particular example is identified by the following iDigBio GUID: | ||
<code> | |||
"idigbio:uuid" : "354210ae-4aa3-49d2-8a66-78a86b019c7b" | |||
</code> | |||
To retrieve a specimen record from v1 of the API with the above iDigBio UUID, we issue an HTTP "GET" request to the /v1/records/{UUID} endpoint with curl: | |||
<code> | |||
http://api.idigbio.org/v1/records/354210ae-4aa3-49d2-8a66-78a86b019c7b | |||
</code> | |||
and receive the following JSON document from the API (in this case, formatted for readability): | |||
<pre> | <pre> | ||
{ | { | ||
"idigbio:uuid" : " | "idigbio:uuid" : "354210ae-4aa3-49d2-8a66-78a86b019c7b", | ||
"idigbio:etag" : " | "idigbio:etag" : "02736fd7318eafed62a4a5ff35175a27fa63983e", | ||
"idigbio:links" : { | "idigbio:links" : { | ||
"mediarecord" : [ | "mediarecord" : [ | ||
"http://api.idigbio.org/v1/mediarecords/ | "http://api.idigbio.org/v1/mediarecords/59141135-813a-4db1-a527-009ae6d17101" | ||
], | ], | ||
"owner" : [ | "owner" : [ | ||
| Line 114: | Line 168: | ||
], | ], | ||
"recordset" : [ | "recordset" : [ | ||
"http://api.idigbio.org/v1/recordsets/ | "http://api.idigbio.org/v1/recordsets/69037495-438d-4dba-bf0f-4878073766f1" | ||
] | ] | ||
}, | }, | ||
"idigbio:version" : | "idigbio:version" : 2, | ||
"idigbio:createdBy" : "872733a2-67a3-4c54-aa76-862735a5f334", | "idigbio:createdBy" : "872733a2-67a3-4c54-aa76-862735a5f334", | ||
"idigbio:recordIds" : [ | "idigbio:recordIds" : [ | ||
"urn:uuid: | "urn:uuid:b036a012-ba1e-41e0-a39a-76fc253640cf" | ||
], | ], | ||
"idigbio:dateModified" : "2014-04- | "idigbio:dateModified" : "2014-04-22T07:33:16.129Z", | ||
"idigbio:data" : { | "idigbio:data" : { | ||
"dwc:catalogNumber" : " | "dwc:day" : "16", | ||
"dwc:identifiedBy" : "CPMAB", | |||
"idigbio:recordId" : "urn:uuid:b036a012-ba1e-41e0-a39a-76fc253640cf", | |||
"dwc:catalogNumber" : "NAUF4A0013309", | |||
"dwc:locality" : "Box Cyn. Santa Rita Mts.", | |||
"dwc:occurrenceID" : "1063507", | |||
"dwc:year" : "1967", | |||
"dwc:recordedBy" : "C.D. Johnson", | |||
"dwc:scientificName" : "Curculionidae", | |||
"dwc:basisOfRecord" : "PreservedSpecimen", | "dwc:basisOfRecord" : "PreservedSpecimen", | ||
"symbiotaverbatimScientificName" : " | "dwc:family" : "Curculionidae", | ||
"dwc:collectionCode" : " | "symbiotaverbatimScientificName" : "Curculionidae", | ||
"dwc: | "dwc:collectionCode" : "NAUF", | ||
"dcterms:references" : "http:// | "dcterms:modified" : "2013-12-20 13:00:36", | ||
"dwc:scientificNameAuthorship" : " | "dwc:country" : "USA", | ||
"dwc:collectionID" : "urn:uuid: | "dcterms:references" : "http://symbiota4.acis.ufl.edu/scan/portal/collections/individual/index.php?occid=1063507", | ||
"dwc:minimumElevationInMeters" : " | "dwc:eventDate" : "1967-08-16", | ||
"dwc: | "dwc:scientificNameAuthorship" : "Latreille, 1802", | ||
"dwc: | "dwc:collectionID" : "urn:uuid:c87a0756-fdd7-4cb6-9921-ca5774f8330e", | ||
"dwc:minimumElevationInMeters" : "1524", | |||
"dwc: | "dwc:verbatimElevation" : "5000'", | ||
"dwc:startDayOfYear" : "228", | |||
"dwc:month" : "8", | |||
"dwc:rights" : "http://creativecommons.org/licenses/by-nc-sa/3.0/", | "dwc:rights" : "http://creativecommons.org/licenses/by-nc-sa/3.0/", | ||
"dwc:stateProvince" : "Arizona", | |||
"dwc:genus" : "Curculionidae", | |||
"dwc:institutionCode" : "NAU", | |||
"dwc:county" : "Pima" | |||
"dwc:stateProvince" : " | |||
"dwc:genus" : " | |||
"dwc: | |||
"dwc: | |||
} | } | ||
} | } | ||
</pre> | </pre> | ||
==== | ==== Elasticsearch ==== | ||
The normalized and indexed record that is stored in Elasticsearch can be retrieved by referencing the iDigBio UUID ("uuid") as the term provided to the query string parameter. | |||
Note that under normal circumstances if one has the iDigBio UUID then there is no reason to use Elasticsearch; the full record is available at the API endpoint. | |||
The list of available terms in the iDigBio index is available at [[IDigBio_API#Elasticsearch_-_Records]]. | The list of available terms in the iDigBio index is available at [[IDigBio_API#Elasticsearch_-_Records]]. | ||
| Line 173: | Line 220: | ||
===== Use the Elasticsearch URI Search ===== | ===== Use the Elasticsearch URI Search ===== | ||
The following command sends the query as an HTTP | The following command sends the query as an HTTP "GET" request: | ||
<pre> | <pre> | ||
| Line 317: | Line 364: | ||
</pre> | </pre> | ||
===== Elasticsearch Request Body Search ===== | ===== Use the Elasticsearch Request Body Search ===== | ||
A JSON query document can be specified on the command-line. Again we are searching for the iDigBio | A JSON query document can be specified on the command-line. Again we are searching for the iDigBio UUID in the "uuid" term. | ||
The following command sends the query as an HTTP | The following command sends the query as an HTTP "POST" request: | ||
<pre> | <pre> | ||
| Line 333: | Line 380: | ||
</pre> | </pre> | ||
and | and gives the following result: | ||
<pre> | <pre> | ||
| Line 476: | Line 523: | ||
</pre> | </pre> | ||
Alternatively, the JSON-formatted query body | Alternatively, the JSON-formatted query body may be sourced from a file. In this example, the file is named uuid.json and contains the iDigBio UUID in the "uuid" term as before: | ||
<pre> | <pre> | ||
| Line 634: | Line 681: | ||
</pre> | </pre> | ||
=== Fetch an Older Version of a Particular | === Fetch an Older Version of a Particular Record === | ||
Consider an iDigBio Media Record "idigbio:uuid" : "ff5fd841-6bc6-4241-80d3-def7e1b6d5e8" | Consider an iDigBio Media Record that is identified by: | ||
<pre> | |||
"idigbio:uuid" : "ff5fd841-6bc6-4241-80d3-def7e1b6d5e8" | |||
</pre> | |||
This record has been updated over time so there are multiple revisions of the record preserved in the API. We can find that the most recent (e.g. "current") version of this particular record is version 1: | |||
<pre> | <pre> | ||
| Line 643: | Line 696: | ||
</pre> | </pre> | ||
We would like to look at the previous version (version 0) to see what has changed. | We would like to look at the previous version (version 0) to see what has changed. We do this by adding the "version" parameter to the url: | ||
<pre> | <pre> | ||
curl -s http://api.idigbio.org/v1/mediarecords/ff5fd841-6bc6-4241-80d3-def7e1b6d5e8?version=0 | json_pp | grep version | |||
"idigbio:version" : "0", | "idigbio:version" : "0", | ||
</pre> | </pre> | ||
The "version" parameter is available for any most of the endpoints that return a single entity record (as opposed to a List). | |||
=== Paging through the API === | |||
Endpoints that return a list of entities may have more total items than are able to be "fit" in a single request. For example, trying to fetch the list of all records in iDigBio will surely timeout in the browser before it is completed (and would put undue load on the server infrastructure). | |||
Request 5 media record entities: | |||
<pre> | <pre> | ||
$ curl -s "http://api.idigbio.org/v1/mediarecords?limit=5" | json_pp | |||
{ | { | ||
"idigbio:errors" : [], | "idigbio:errors" : [], | ||
| Line 714: | Line 734: | ||
}, | }, | ||
"idigbio:uuid" : "00000728-ffb3-4a68-9f93-137f19961121", | "idigbio:uuid" : "00000728-ffb3-4a68-9f93-137f19961121", | ||
"idigbio:version" : | "idigbio:version" : 3, | ||
"idigbio:etag" : " | "idigbio:etag" : "ef2cac326a60d89d8cb9005abaa82068bfa83565", | ||
"idigbio:dateModified" : "2014-04- | "idigbio:dateModified" : "2014-04-24T05:03:56.782Z" | ||
}, | }, | ||
{ | { | ||
| Line 741: | Line 761: | ||
}, | }, | ||
"idigbio:uuid" : "000012f9-d288-4a14-b898-77430e0a137a", | "idigbio:uuid" : "000012f9-d288-4a14-b898-77430e0a137a", | ||
"idigbio:version" : 1, | |||
"idigbio:etag" : "cf49416750fdb9bdb808c334a74b84f27bb8160b", | |||
"idigbio:dateModified" : "2014-04-23T02:43:08.344Z" | |||
} | |||
], | |||
"idigbio:itemCount" : "2342880" | |||
} | |||
</pre> | |||
The next "page" of records can be requested by adding the "offset" paramenter. | |||
A link that includes the offset to the next page is provided in the results: | |||
<pre> | |||
"idigbio:links" : { | |||
"idigbio:nextPage" : "http://api.idigbio.org/v1/mediarecords?limit=5&offset=5" | |||
} | |||
</pre> | |||
Request the next "page" of entity ids by starting at offset 5. | |||
<pre> | |||
$ curl -s "http://api.idigbio.org/v1/mediarecords?limit=5&offset=5" | json_pp | |||
{ | |||
"idigbio:errors" : [], | |||
"idigbio:links" : { | |||
"idigbio:nextPage" : "http://api.idigbio.org/v1/mediarecords?limit=5&offset=10", | |||
"idigbio:prevPage" : "http://api.idigbio.org/v1/mediarecords?limit=5&offset=0" | |||
}, | |||
"idigbio:items" : [ | |||
{ | |||
"idigbio:links" : { | |||
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/00001478-c150-4faf-a617-439a838d4377" | |||
}, | |||
"idigbio:uuid" : "00001478-c150-4faf-a617-439a838d4377", | |||
"idigbio:version" : 1, | |||
"idigbio:etag" : "30f602e4eb47ebb2ceb265f64217e3cf5664f517", | |||
"idigbio:dateModified" : "2014-03-21T23:09:39.752Z" | |||
}, | |||
{ | |||
"idigbio:links" : { | |||
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/00001a91-189b-4002-b56e-a770a55951a0" | |||
}, | |||
"idigbio:uuid" : "00001a91-189b-4002-b56e-a770a55951a0", | |||
"idigbio:version" : 0, | |||
"idigbio:etag" : "647e82d17ee435fb14f0f8607dabe88dfc3a1944", | |||
"idigbio:dateModified" : "2014-04-25T04:49:32.359Z" | |||
}, | |||
{ | |||
"idigbio:links" : { | |||
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/00002091-4fb3-410a-9307-bd3e917dfcca" | |||
}, | |||
"idigbio:uuid" : "00002091-4fb3-410a-9307-bd3e917dfcca", | |||
"idigbio:version" : 0, | |||
"idigbio:etag" : "90d98d48d9e7e07eab9064bd9b6e22ce6502c07f", | |||
"idigbio:dateModified" : "2014-05-03T18:45:47.112Z" | |||
}, | |||
{ | |||
"idigbio:links" : { | |||
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/00002c32-ae3a-41ed-9bd9-f6c50d3e35fb" | |||
}, | |||
"idigbio:uuid" : "00002c32-ae3a-41ed-9bd9-f6c50d3e35fb", | |||
"idigbio:version" : 3, | |||
"idigbio:etag" : "d1ded90d06e93876b1badd01222905add93e8806", | |||
"idigbio:dateModified" : "2014-04-19T00:25:59.471Z" | |||
}, | |||
{ | |||
"idigbio:links" : { | |||
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/00002dbd-6415-463b-8cae-38f548415ffa" | |||
}, | |||
"idigbio:uuid" : "00002dbd-6415-463b-8cae-38f548415ffa", | |||
"idigbio:version" : 2, | |||
"idigbio:etag" : "4e298045b496146f5c51e331c9887fd7afde4deb", | |||
"idigbio:dateModified" : "2014-04-21T20:29:39.531Z" | |||
} | |||
], | |||
"idigbio:itemCount" : "2342880" | |||
} | |||
</pre> | |||
which includes links to the previous page and next page: | |||
<pre> | |||
"idigbio:nextPage" : "http://api.idigbio.org/v1/mediarecords?limit=5&offset=10", | |||
"idigbio:prevPage" : "http://api.idigbio.org/v1/mediarecords?limit=5&offset=0" | |||
</pre> | |||
With current results starting at offset 5 and a limit of 5, the previous page and a next page offsets are 0 and 10 respectively (offset +- limit). | |||
;TO DO One more paging to help differentiate these multiples of 5. | |||
=== Fetch a list of five MediaRecord IDs === | |||
The "mediarecords" endpoint returns a List of Media Record IDs. | |||
Example URL: | |||
http://api.idigbio.org/v1/mediarecords?limit=5&offset=5 | |||
Example Results: | |||
<pre> | |||
$ curl -s "http://api.idigbio.org/v1/mediarecords?limit=5&offset=5" | json_pp | |||
{ | |||
"idigbio:errors" : [], | |||
"idigbio:links" : { | |||
"idigbio:nextPage" : "http://api.idigbio.org/v1/mediarecords?limit=5&offset=10", | |||
"idigbio:prevPage" : "http://api.idigbio.org/v1/mediarecords?limit=5&offset=0" | |||
}, | |||
"idigbio:items" : [ | |||
{ | |||
"idigbio:links" : { | |||
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/00001478-c150-4faf-a617-439a838d4377" | |||
}, | |||
"idigbio:uuid" : "00001478-c150-4faf-a617-439a838d4377", | |||
"idigbio:version" : 1, | |||
"idigbio:etag" : "30f602e4eb47ebb2ceb265f64217e3cf5664f517", | |||
"idigbio:dateModified" : "2014-03-21T23:09:39.752Z" | |||
}, | |||
{ | |||
"idigbio:links" : { | |||
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/00001a91-189b-4002-b56e-a770a55951a0" | |||
}, | |||
"idigbio:uuid" : "00001a91-189b-4002-b56e-a770a55951a0", | |||
"idigbio:version" : 0, | |||
"idigbio:etag" : "647e82d17ee435fb14f0f8607dabe88dfc3a1944", | |||
"idigbio:dateModified" : "2014-04-25T04:49:32.359Z" | |||
}, | |||
{ | |||
"idigbio:links" : { | |||
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/00002091-4fb3-410a-9307-bd3e917dfcca" | |||
}, | |||
"idigbio:uuid" : "00002091-4fb3-410a-9307-bd3e917dfcca", | |||
"idigbio:version" : 0, | "idigbio:version" : 0, | ||
"idigbio:etag" : " | "idigbio:etag" : "90d98d48d9e7e07eab9064bd9b6e22ce6502c07f", | ||
"idigbio:dateModified" : "2014- | "idigbio:dateModified" : "2014-05-03T18:45:47.112Z" | ||
}, | |||
{ | |||
"idigbio:links" : { | |||
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/00002c32-ae3a-41ed-9bd9-f6c50d3e35fb" | |||
}, | |||
"idigbio:uuid" : "00002c32-ae3a-41ed-9bd9-f6c50d3e35fb", | |||
"idigbio:version" : 3, | |||
"idigbio:etag" : "d1ded90d06e93876b1badd01222905add93e8806", | |||
"idigbio:dateModified" : "2014-04-19T00:25:59.471Z" | |||
}, | |||
{ | |||
"idigbio:links" : { | |||
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/00002dbd-6415-463b-8cae-38f548415ffa" | |||
}, | |||
"idigbio:uuid" : "00002dbd-6415-463b-8cae-38f548415ffa", | |||
"idigbio:version" : 2, | |||
"idigbio:etag" : "4e298045b496146f5c51e331c9887fd7afde4deb", | |||
"idigbio:dateModified" : "2014-04-21T20:29:39.531Z" | |||
} | } | ||
], | ], | ||
"idigbio:itemCount" : " | "idigbio:itemCount" : "2342880" | ||
} | } | ||
</pre> | </pre> | ||
Screenshot of Results: | |||
[[File:Screenshot_jsonview_idigbio_api_mediarecords_limit_offset.png]] | |||
=== Display a thumbnail image for a specimen record === | |||
Let's say that we wish to display a thumbnail image for a particular specimen record. | |||
We can look at the full record: | |||
<pre> | |||
$ curl -s "http://api.idigbio.org/v1/records/182c9260-0151-454a-a648-4d58ddf51bd8" | json_pp | |||
{ | |||
"idigbio:uuid" : "182c9260-0151-454a-a648-4d58ddf51bd8", | |||
"idigbio:etag" : "bb5ee46e2730948c946c178e8da85dedee4bea80", | |||
"idigbio:links" : { | |||
"mediarecord" : [ | |||
"http://api.idigbio.org/v1/mediarecords/00002091-4fb3-410a-9307-bd3e917dfcca" | |||
], | |||
"owner" : [ | |||
"872733a2-67a3-4c54-aa76-862735a5f334" | |||
], | |||
"recordset" : [ | |||
"http://api.idigbio.org/v1/recordsets/09edf7d2-e68e-4a42-93da-762f86bb814f" | |||
] | |||
}, | |||
"idigbio:version" : 0, | |||
"idigbio:createdBy" : "872733a2-67a3-4c54-aa76-862735a5f334", | |||
"idigbio:recordIds" : [ | |||
"urn:uuid:7d0d4400-7c6c-4bb4-b2ad-7b1f6e776755" | |||
], | |||
"idigbio:dateModified" : "2014-05-03T14:27:40.596Z", | |||
"idigbio:data" : { | |||
"idigbio:recordId" : "urn:uuid:7d0d4400-7c6c-4bb4-b2ad-7b1f6e776755", | |||
"dwc:catalogNumber" : "CBS.038533", | |||
"dwc:specificEpithet" : "novae-angliae", | |||
"dwc:occurrenceID" : "272024", | |||
"dwc:year" : "0", | |||
"dwc:recordedBy" : "Hugh S. Clark", | |||
"dwc:scientificName" : "Symphyotrichum novae-angliae", | |||
"dwc:basisOfRecord" : "PreservedSpecimen", | |||
"dwc:family" : "Asteraceae", | |||
"symbiotaverbatimScientificName" : "Symphyotrichum novae-angliae", | |||
"dwc:collectionCode" : "CBS", | |||
"dcterms:modified" : "2012-03-14 00:00:00", | |||
"dwc:decimalLongitude" : "-72.6834", | |||
"dwc:country" : "USA", | |||
"dcterms:references" : "http://portal.neherbaria.org/portal/collections/individual/index.php?occid=272024", | |||
"dwc:municipality" : "Hartford", | |||
"dwc:scientificNameAuthorship" : "(L.) G.L. Nesom", | |||
"dwc:month" : "0", | |||
"dwc:rights" : "http://hdl.handle.net/10079/8931zqj", | |||
"dwc:geodeticDatum" : "WGS84", | |||
"dwc:rightsHolder" : "Peabody Museum of Natural History, Yale University", | |||
"dwc:stateProvince" : "Connecticut", | |||
"dwc:genus" : "Symphyotrichum", | |||
"dwc:coordinateUncertaintyInMeters" : "5419", | |||
"dwc:decimalLatitude" : "41.766", | |||
"dwc:accessRights" : "http://hdl.handle.net/10079/8931zqj", | |||
"dwc:institutionCode" : "YPM", | |||
"dwc:county" : "Hartford County" | |||
} | |||
} | |||
</pre> | |||
At this point we have a couple of options to get to the thumbnail. The easiest is to use the /media endpoint for this record's iDigBio UUID and add the "quality" parameter with "thumbnail" as the value: | |||
http://api.idigbio.org/v1/records/182c9260-0151-454a-a648-4d58ddf51bd8/media?quality=thumbnail | |||
which results in a number of HTTP Redirects to the target thumbnail image, illustrated here with the Chrome Developer Tools visible: | |||
[[File:Screenshot_firefox_webdevtools_thumbnail_specimen_182c9260-0151-454a-a648-4d58ddf51bd8.png]] | |||
''Note that the final image URL should not be considered stable or permanent. Images can be moved around to various storage platforms. The Mediarecord has a UUID that will remain contstant but the containing data, including the image location, may be updated over time.'' | |||
The /media endpoint of a specimen record is kind of shortcut to avoid having to walk through the relationship to a mediarecord and then to an image file. | |||
Alternatively, if we actually wanted to look at media metadata, we could use the link to the mediarecord in the <code>"idigbio:links"</code> section: | |||
<pre> | |||
"idigbio:links" : { | |||
"mediarecord" : [ | |||
"http://api.idigbio.org/v1/mediarecords/00002091-4fb3-410a-9307-bd3e917dfcca" | |||
], | |||
</pre> | |||
which also has a /media endpoint that can lead to the same thumbnail image. | |||
http://api.idigbio.org/v1/mediarecords/00002091-4fb3-410a-9307-bd3e917dfcca/media?quality=thumbnail | |||
=== Advanced Elasticsearch Examples === | === Advanced Elasticsearch Examples === | ||
; 1. Find all specimen records that meet the following | {{Caution|'''Direct queries to the iDigBio Elasticsearch service are deprecated. The [https://github.com/idigbio/idigbio-search-api/wiki iDigBio API v2 / iDigBio Search API specification] should be used instead.'''}} | ||
==== Search using Locality Data ==== | |||
The [[IDigBio API v1 Specification#Search|iDigBio Elasticsearch]] documentation should be considered required reading for these advanced Elasticsearch examples. | |||
; 1. Find all specimen records that meet the following criteria: | |||
* Has an image (mediarecord) associated with the specimen record | * Has an image (mediarecord) associated with the specimen record | ||
| Line 774: | Line 1,033: | ||
# "hasImage" is true | # "hasImage" is true | ||
# "county" is either "Hocking" or "Fairfield" | # "county" is either "Hocking" or "Fairfield" | ||
# "stateprovince" is | # "stateprovince" is "Ohio" (there are multiple Fairfield counties throughout the world so we need to restrict the geography by state) | ||
The essential query parameter string will look something like this: | The essential query parameter string will look something like this (note we must lowercase the values): | ||
<pre> | <pre> | ||
| Line 782: | Line 1,041: | ||
</pre> | </pre> | ||
In addition, it is possible to request more or fewer records at a time using the "from" and "size" parameters. Below is the full curl example requesting only the first 3 records and grep | In addition, it is possible to request more or fewer records at a time using the "from" and "size" parameters. Below is the full curl example requesting only the first 3 records and using grep on the output to filter everything except the iDigBio UUID field. Note that we quote the full URL to keep the shell from trying to interpret the special characters. | ||
<pre> | <pre> | ||
$ curl -s | $ curl -s 'http://search.idigbio.org/idigbio/records/_search?from=0&size=3&q=hasImage:true+AND+county:(fairfield+OR+hocking)+AND+stateprovince:ohio' | json_pp | egrep "\"uuid\"" | ||
"uuid" : "ecf14f53-d225-4eb5-89e6-66379b360781", | |||
"uuid" : "e45071cd-40eb-41e7-9105-0e35234025f5", | |||
"uuid" : "20ad60a8-4126-4cc0-9378-411a9a640005", | "uuid" : "20ad60a8-4126-4cc0-9378-411a9a640005", | ||
</pre> | </pre> | ||
Now that we have iDigBio UUIDs, we could retrieve images for these specimens by iterating over the /media endpoints for each of those uuids in the API. | |||
http://api.idigbio.org/v1/records/ecf14f53-d225-4eb5-89e6-66379b360781/media?quality=webview | |||
http://api.idigbio.org/v1/records/e45071cd-40eb-41e7-9105-0e35234025f5/media?quality=webview | |||
... | |||
Note that we did not specify any sorting in the Elasticsearch query and we did restrict the result count by using the "size" parameter. If the same query is run at a later time the specific records returned could be different. | |||
Note also that the above query is for an exact match on the county name which will perform fairly well. Unfortunately, the data includes many variations in the county field for these counties such as: | |||
<pre> | |||
"Fairfield" | |||
"Fairfield County" | |||
"Hocking" | |||
"Hocking Co." | |||
"Hocking County" | |||
"Hocking Hills" | |||
</pre> | |||
To find these related records, a wildcard query may be useful. In the following case the size is also increased to 3000 to retrieve a larger sample. | |||
<pre> | <pre> | ||
$ curl -s 'http://search.idigbio.org/idigbio/records/_search?from=0&size=3000&q=hasImage:true+AND+county:(fairfield*+OR+hocking*)+AND+stateprovince:ohio' | json_pp | |||
</pre> | </pre> | ||
==== Family: Curculionidae - Search Example ==== | |||
{{Caution|'''Direct queries to the iDigBio Elasticsearch service are deprecated. The [https://github.com/idigbio/idigbio-search-api/wiki iDigBio API v2 / iDigBio Search API specification] should be used instead.'''}} | |||
Let us say we are looking for images associated with specimen records for Family Curculionidae (a family of weevils). | |||
If we already know the iDigBio UUID of a specimen we can simply use the <code>/media</code> endpoint for the record: | |||
<code> | |||
http://api.idigbio.org/v1/records/c2e34bda-0405-48ea-b7b2-76f893777f20/media?quality=webview | |||
</code> | |||
Giving us an image from the iDigBio API that looks something like this: | |||
[[File:Curculionidae_example_Acallodes_ventricosus.jpg]] | |||
But how did we find the proper iDigBio UUID for this specimen record in the first place? | |||
The [https://www.idigbio.org/portal/search iDigBio Portal Search] is of course built just for this purpose. | |||
If on the other hand we wanted to search for these specimens via a software program, we could use the iDigBio Elasticsearch interface. | |||
I happen to know that this specimen was collected in New Jersey, so we search for records of Family Curculionidae in New Jersey that have associated images. Note the special encoding of the query string (space becomes %20, quotation becomes %22) required when using curl vs. a browser (the browser would do this encoding for you). | |||
'''''The following example may not be correct yet...''''' | |||
<pre>$ curl -s 'http://search.idigbio.org/idigbio/records/_search?q=hasImage:true+AND+family:curculionidae+AND+stateprovince:%22new%20jersey%22'</pre> | |||
=== Mapping Example === | === Mapping Example === | ||
* | {{Caution|'''Direct queries to the iDigBio Elasticsearch service are deprecated. The [https://github.com/idigbio/idigbio-search-api/wiki iDigBio API v2 / iDigBio Search API specification] should be used instead.'''}} | ||
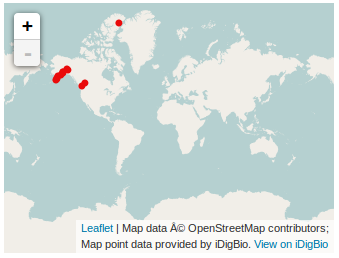
This Mapping example is really just another Elasticsearch example. To generate a map, we basically want to do an Elasticsearch query to choose our specimens of interest and then put the points for those specimens on a map. | |||
The [[IDigBio API v1 Specification#Search|iDigBio Elasticsearch]] documentation should be considered required reading before attempting to integrate this code into one's site. | |||
This example displays a map that includes specimen data points that match the criteria of Scientific Name (dwc:scientificName) is "Abietinella abietina" and Institution Code (dwc:institutionCode) is "DUKE". | |||
The Elasticsearch terms for this query are: | |||
<pre> | |||
scientificname" : "abietinella abietina" | |||
institutioncode": "duke" | |||
</pre> | |||
The following is a full HTML page with javascript code that displays a small specimen map containing iDigBio data points. | |||
To customize the map search results, modify the <code>var terms = [ ... </code> section with the terms of your choice. | |||
<pre> | |||
<!DOCTYPE html> | |||
<html lang="en"> | |||
<head> | |||
<title>iDigBio Map</title> | |||
<link rel="stylesheet" href="http://cdn.leafletjs.com/leaflet-0.7.2/leaflet.css" /> | |||
<script src="http://code.jquery.com/jquery-1.11.0.min.js"></script> | |||
<script src="http://cdn.leafletjs.com/leaflet-0.7.2/leaflet.js"></script> | |||
<script type="text/javascript"> | |||
/* | |||
* This example provided by iDigBio. www.idigbio.org | |||
* | |||
* The list of available terms is available at | |||
* https://www.idigbio.org/wiki/index.php/IDigBio_API_v1_Specification#Search | |||
* | |||
* Some Example Terms: | |||
* uuid, scientificname, institutioncode, collectioncode, genus, | |||
* country, stateprovince, county, occurenceid, barcodevalue, catalognumber | |||
*****/ | |||
var terms = [ | |||
{"term" : { "scientificname" : "abietinella abietina"}}, | |||
{"term" : { "institutioncode": "duke"}} | |||
]; | |||
$(document).ready(function(){ | |||
var query = { | |||
"size": 10000, | |||
"from": 0, | |||
"query":{ | |||
"filtered":{ | |||
"filter":{ | |||
"and":[] | |||
} | |||
} | |||
}, | |||
"sort":{ | |||
"scientificname":{"order":"asc"} | |||
} | |||
}; | |||
/* | |||
*Build query params and outlink | |||
***/ | |||
var link=[]; | |||
$.each(terms,function(ind,item){ | |||
query.query.filtered.filter.and.push(item); | |||
$.each(item.term,function(key,val){ | |||
link.push(key+'='+ val.toLowerCase()); | |||
}); | |||
}); | |||
var link_href = 'https://www.idigbio.org/portal/search?'+link.join('&'); | |||
var link_string = '<br>Map point data provided by iDigBio. <a id="link" href="' + link_href + '">View on iDigBio</a>'; | |||
var base = L.tileLayer('http://{s}.tile.openstreetmap.org/{z}/{x}/{y}.png',{ | |||
attribution: 'Map data © OpenStreetMap contributors; ' + link_string, | |||
minZoom: 0, | |||
maxZoom: 18, | |||
reuseTiles: true | |||
}); | |||
map = L.map('map',{ | |||
center: [0,0], | |||
zoom: 0, | |||
layers: [base], | |||
scrollWheelZoom: true, | |||
boxZoom: false | |||
}); | |||
/* | |||
*Make request to iDigBio search server | |||
***/ | |||
$.post('https://search.idigbio.org/idigbio/records/_search', JSON.stringify(query),function(resp){ | |||
$(resp.hits.hits).each(function(ind,item){ | |||
var source = item._source; | |||
if(typeof source.geopoint !== 'undefined'){ | |||
var c = new L.Circle([source.geopoint.lat,source.geopoint.lon],1,{color:'#E90A0A',opacity:1,fillopacity:1}); | |||
c.addTo(map); | |||
} | |||
}); | |||
}); | |||
}); | |||
</script> | |||
</head> | |||
<body> | |||
<div id="map" style="height:250px;width:330px;"></div> | |||
</body> | |||
</html> | |||
</pre> | |||
The above html page would render as follows (captured in a screenshot) in a browser: | |||
[[File:Idigbio_api_mapping_search_example.png]] | |||
Deeper integration of iDigBio mapping requires understanding of geohashes and would be a more significant undertaking. If your project wishes advanced integration, please contact iDigBio. https://www.idigbio.org/contact | |||
=== idigbio:links and the 20 Item List === | |||
Compare the idigbio:links section of the following two Publisher records: | |||
http://api.idigbio.org/v1/publishers/076c0ff6-65e9-48a5-8e4b-2447936f9a1c | |||
http://api.idigbio.org/v1/publishers/4e1beef9-d7c0-4ac0-87df-065bc5a55361 | |||
The first Publisher record returns a JSON document containing "idigbio:links" and a List of recordsets (or to be more precise, a List of API urls that can be used to access individual Recordset records by iDigBio UUID). Since the number of recordsets is less than 20, they are included in the Publisher record output. | |||
<pre> | |||
$ curl -s http://api.idigbio.org/v1/publishers/076c0ff6-65e9-48a5-8e4b-2447936f9a1c | json_pp | |||
{ | |||
"idigbio:uuid" : "076c0ff6-65e9-48a5-8e4b-2447936f9a1c", | |||
"idigbio:etag" : "31b115d5a307027f757554441c93d8e5b8ac8fab", | |||
"idigbio:links" : { | |||
"recordset" : [ | |||
"http://api.idigbio.org/v1/recordsets/197f7bb8-213d-4fae-b536-652dd91e56dc", | |||
"http://api.idigbio.org/v1/recordsets/31c140bc-e6f1-4acc-beaf-b825cf288ad9", | |||
"http://api.idigbio.org/v1/recordsets/5aac25d2-bcfb-4084-a700-584311ea539d", | |||
"http://api.idigbio.org/v1/recordsets/5e893602-84ca-4c8c-bac1-99111c777582", | |||
"http://api.idigbio.org/v1/recordsets/6539877e-82dc-485c-ad3d-038f383d5431", | |||
"http://api.idigbio.org/v1/recordsets/69037495-438d-4dba-bf0f-4878073766f1", | |||
"http://api.idigbio.org/v1/recordsets/7fcdca8e-7469-480c-8516-cce4e24c37c9", | |||
"http://api.idigbio.org/v1/recordsets/82541f90-fe8e-4d66-84d8-4fe515dc5533", | |||
"http://api.idigbio.org/v1/recordsets/89eb1ad0-ae60-4e8a-bf34-a53d0423bc80", | |||
"http://api.idigbio.org/v1/recordsets/92dd8c8e-c048-4f0a-9b5d-2ee627d2f553", | |||
"http://api.idigbio.org/v1/recordsets/9ace4c05-d930-45c7-8d2d-0cadff1ea32b", | |||
"http://api.idigbio.org/v1/recordsets/9bd4ff72-1cb2-431f-bf7b-b5d47e08cc02", | |||
"http://api.idigbio.org/v1/recordsets/a3b77120-3770-46dd-ba47-6941eff848b3", | |||
"http://api.idigbio.org/v1/recordsets/ab4b6a2b-a90a-44ce-95a1-2c44c911fcc6", | |||
"http://api.idigbio.org/v1/recordsets/cb65cf5e-07b8-4d53-a91a-7dce9b8ccf80", | |||
"http://api.idigbio.org/v1/recordsets/d315c4a3-0bee-49d1-8d03-726358937cde", | |||
"http://api.idigbio.org/v1/recordsets/d767f759-af64-4464-8614-c77ca44cad8d", | |||
"http://api.idigbio.org/v1/recordsets/f83517ea-7b9f-443d-9371-d11a05ebc0a7", | |||
"http://api.idigbio.org/v1/recordsets/ff111763-e72d-4f24-8914-b5b2dd94908c" | |||
] | |||
}, | |||
... | |||
</pre> | |||
The second link returns a JSON document containing "idigbio:links" and a single recordset link rather than a List. To get to the List of recordsets for this Publisher record, this intermediate link needs to be followed. | |||
<pre> | |||
$ curl -s http://api.idigbio.org/v1/publishers/4e1beef9-d7c0-4ac0-87df-065bc5a55361 | json_pp | |||
{ | |||
"idigbio:uuid" : "4e1beef9-d7c0-4ac0-87df-065bc5a55361", | |||
"idigbio:etag" : "01c8eabbf9e8fe7f5af86349b04cad55b3ddee8e", | |||
"idigbio:links" : { | |||
"recordset" : [ | |||
"http://api.idigbio.org/v1/publishers/4e1beef9-d7c0-4ac0-87df-065bc5a55361/recordsets" | |||
] | |||
}, | |||
... | |||
</pre> | |||
As suspected, the list of Recordsets includes more than 20 items (in this case, 51 recordset items) which is why they were not displayed with the Publisher record. | |||
<pre> | |||
$ curl -s http://api.idigbio.org/v1/publishers/4e1beef9-d7c0-4ac0-87df-065bc5a55361/recordsets | json_pp | grep recordset | wc -l | |||
51 | |||
$ curl -s http://api.idigbio.org/v1/publishers/4e1beef9-d7c0-4ac0-87df-065bc5a55361/recordsets | json_pp | |||
{ | |||
"idigbio:errors" : [], | |||
"idigbio:links" : {}, | |||
"idigbio:items" : [ | |||
{ | |||
"idigbio:links" : { | |||
"recordset" : "http://api.idigbio.org/v1/recordsets/0072bf11-a354-4998-8730-c0cb4cfc9517" | |||
}, | |||
"idigbio:uuid" : "0072bf11-a354-4998-8730-c0cb4cfc9517" | |||
}, | |||
{ | |||
"idigbio:links" : { | |||
"recordset" : "http://api.idigbio.org/v1/recordsets/00d9fcc1-c8e2-4ef6-be64-9994ca6a32c3" | |||
}, | |||
"idigbio:uuid" : "00d9fcc1-c8e2-4ef6-be64-9994ca6a32c3" | |||
}, | |||
{ | |||
"idigbio:links" : { | |||
"recordset" : "http://api.idigbio.org/v1/recordsets/0dd7c8dd-412c-4a13-a1d3-47e1e1af5455" | |||
}, | |||
"idigbio:uuid" : "0dd7c8dd-412c-4a13-a1d3-47e1e1af5455" | |||
}, | |||
{ | |||
"idigbio:links" : { | |||
"recordset" : "http://api.idigbio.org/v1/recordsets/181352ea-3598-4f32-b919-c8f6097f4c65" | |||
}, | |||
"idigbio:uuid" : "181352ea-3598-4f32-b919-c8f6097f4c65" | |||
}, | |||
{ | |||
"idigbio:links" : { | |||
"recordset" : "http://api.idigbio.org/v1/recordsets/196bb137-2f82-40d5-b294-e730af29749f" | |||
}, | |||
"idigbio:uuid" : "196bb137-2f82-40d5-b294-e730af29749f" | |||
}, | |||
... | |||
</pre> | |||
=== API Performance === | === API Performance === | ||
Latest revision as of 18:09, 9 July 2018
| The v1 API has been superseded by iDigBio API v2 / iDigBio Search API. API users should migrate to using the most recent version of the API. This page remains for historical reference. |
This document is a list of examples for using the IDigBio API but may not be comprehensive to cover every feature of the API. See iDigBio API v1 Specification for full documentation on the API including all of the available endpoints and parameters.
Viewing JSON Output
Most of the examples provided here will be shown using the curl command-line tool. The dollar sign $ is an indication of a command prompt where something is typed into a shell or terminal. To run the commands, the $ should be omitted.
Here are some of the more commonly used curl options in our examples (for descriptions and additional usage information, see the curl man page):
$ man curl
...
-s, --silent
-v, --verbose
-d, --data <data>
...
Many of the examples will also include "pipes" to chain multiple commands together. The Wikipedia article on Unix software pipelines may be a useful reference.
Javascript Object Notation (JSON) Quick Intro
Javascript Object Notation (JSON), is an open standard format that uses human-readable text to transmit data objects consisting of attribute–value pairs. JSON has become a common format for interchanging data via APIs and especially modern Web Services. Effective use of the iDigBio API will require some understanding of JSON.
A simple JSON document with two fields and two data values would look something like this:
{
"field1" : "value_a",
"field2" : "value_b"
}
If we were storing data about John Smith, age 50, we might represent it as the following JSON document:
{
"lastName" : "Smith",
"firstName" : "John",
"age" : "50"
}
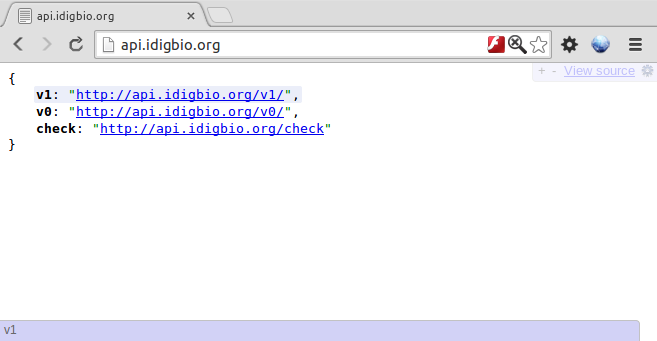
A short JSON document is returned by the iDigBio API when calling the HTTP "GET" method (curl's default) against the base URL (http://api.idigbio.org):
{
"v1" : "http://api.idigbio.org/v1/",
"check" : "http://api.idigbio.org/check",
"v0" : "http://api.idigbio.org/v0/"
}
It may be useful to inspect the API's JSON document outputs using a "pretty printing" feature of some kind. This will make it more readable by humans and also allow it to be more easily filtered using tools such as grep. Two common tools for this available on Unix/Linux are python's json.tool module or the json_pp command-line Perl script (JSON::PP). In typical Unix fashion, these tools can receive input from a pipe and be used to chain multiple commands together.
There are also ways of viewing JSON inside of a web browser.
Most of our examples in this document will include piping the JSON document into json_pp.
JSONView Web Browser Extension
For viewing of JSON results in a browser, one may wish to use the JSONView Extension which is available for Firefox and Chrome.
Here is a screenshot of the output using the JSONView extension after requesting the API base URL:
json.tool (Python)
Current installations of Python typically include a json module in the standard installed libraries.
$ curl -s http://api.idigbio.org/ | python -m json.tool
{
"check": "http://api.idigbio.org/check",
"v0": "http://api.idigbio.org/v0/",
"v1": "http://api.idigbio.org/v1/"
}
json_pp (Perl)
Similarly, modern Perl installations typically include the json_pp script or it can be aquired from CPAN.
$ curl -s http://api.idigbio.org/ | json_pp
{
"v1" : "http://api.idigbio.org/v1/",
"check" : "http://api.idigbio.org/check",
"v0" : "http://api.idigbio.org/v0/"
}
simplejson.tool (Python)
If a Python installation is misssing the json standard library, the separate simplejson package may solve the problem.
The simplejson.tool module can be invoked in the same way as json.tool:
$ curl -s http://api.idigbio.org/ | python -m simplejson.tool
{
"check": "http://api.idigbio.org/check",
"v0": "http://api.idigbio.org/v0/",
"v1": "http://api.idigbio.org/v1/"
}
Older versions of python also may require a more complicated syntax to invoke the module from the command line.
$ curl -s http://api.idigbio.org/ | python -c'from simplejson.tool import main; main()'
{
"check": "http://api.idigbio.org/check",
"v0": "http://api.idigbio.org/v0/",
"v1": "http://api.idigbio.org/v1/"
}
Three ways to fetch a specific record based on iDigBio UUID
The following three examples intend to illustrate the various methods for looking up a unique record in iDigBio. Each record that is added to iDigBio is assigned a Globally Unique Identifier (GUID) of the UUID type.
The data returned by the Elasticsearch interface may differ from the raw API record, especially with regards to field names, since the Elasticsearch data has been normalized and indexed for consumption by the search portal.
Regardless of whether the record's data is retrieved by using the API or by using Elasticsearch, the piece of information used to identify a record is the iDigBio UUID.
For these three examples, consider the specimen record identified by the following iDigBio UUID:
"idigbio:uuid" : "0cec43d8-9ad5-478a-bea1-397ab5cc4430"
Use the API
A complete record that was provided to iDigBio is stored in the API and can be retrieved by referencing the iDigBio UUID ("idigbio:uuid") at the appropriate endpoint.
Let us say that we have already located the specimen record for a particular Curculionidae specimen (a family of weevils). The specimen record for our particular example is identified by the following iDigBio GUID:
"idigbio:uuid" : "354210ae-4aa3-49d2-8a66-78a86b019c7b"
To retrieve a specimen record from v1 of the API with the above iDigBio UUID, we issue an HTTP "GET" request to the /v1/records/{UUID} endpoint with curl:
http://api.idigbio.org/v1/records/354210ae-4aa3-49d2-8a66-78a86b019c7b
and receive the following JSON document from the API (in this case, formatted for readability):
{
"idigbio:uuid" : "354210ae-4aa3-49d2-8a66-78a86b019c7b",
"idigbio:etag" : "02736fd7318eafed62a4a5ff35175a27fa63983e",
"idigbio:links" : {
"mediarecord" : [
"http://api.idigbio.org/v1/mediarecords/59141135-813a-4db1-a527-009ae6d17101"
],
"owner" : [
"872733a2-67a3-4c54-aa76-862735a5f334"
],
"recordset" : [
"http://api.idigbio.org/v1/recordsets/69037495-438d-4dba-bf0f-4878073766f1"
]
},
"idigbio:version" : 2,
"idigbio:createdBy" : "872733a2-67a3-4c54-aa76-862735a5f334",
"idigbio:recordIds" : [
"urn:uuid:b036a012-ba1e-41e0-a39a-76fc253640cf"
],
"idigbio:dateModified" : "2014-04-22T07:33:16.129Z",
"idigbio:data" : {
"dwc:day" : "16",
"dwc:identifiedBy" : "CPMAB",
"idigbio:recordId" : "urn:uuid:b036a012-ba1e-41e0-a39a-76fc253640cf",
"dwc:catalogNumber" : "NAUF4A0013309",
"dwc:locality" : "Box Cyn. Santa Rita Mts.",
"dwc:occurrenceID" : "1063507",
"dwc:year" : "1967",
"dwc:recordedBy" : "C.D. Johnson",
"dwc:scientificName" : "Curculionidae",
"dwc:basisOfRecord" : "PreservedSpecimen",
"dwc:family" : "Curculionidae",
"symbiotaverbatimScientificName" : "Curculionidae",
"dwc:collectionCode" : "NAUF",
"dcterms:modified" : "2013-12-20 13:00:36",
"dwc:country" : "USA",
"dcterms:references" : "http://symbiota4.acis.ufl.edu/scan/portal/collections/individual/index.php?occid=1063507",
"dwc:eventDate" : "1967-08-16",
"dwc:scientificNameAuthorship" : "Latreille, 1802",
"dwc:collectionID" : "urn:uuid:c87a0756-fdd7-4cb6-9921-ca5774f8330e",
"dwc:minimumElevationInMeters" : "1524",
"dwc:verbatimElevation" : "5000'",
"dwc:startDayOfYear" : "228",
"dwc:month" : "8",
"dwc:rights" : "http://creativecommons.org/licenses/by-nc-sa/3.0/",
"dwc:stateProvince" : "Arizona",
"dwc:genus" : "Curculionidae",
"dwc:institutionCode" : "NAU",
"dwc:county" : "Pima"
}
}
Elasticsearch
The normalized and indexed record that is stored in Elasticsearch can be retrieved by referencing the iDigBio UUID ("uuid") as the term provided to the query string parameter.
Note that under normal circumstances if one has the iDigBio UUID then there is no reason to use Elasticsearch; the full record is available at the API endpoint.
The list of available terms in the iDigBio index is available at IDigBio_API#Elasticsearch_-_Records.
Use the Elasticsearch URI Search
The following command sends the query as an HTTP "GET" request:
$ curl -s "https://search.idigbio.org/idigbio/records/_search?q=uuid:0cec43d8-9ad5-478a-bea1-397ab5cc4430" | json_pp
{
"hits" : {
"hits" : [
{
"_source" : {
"collectioncode" : "plants",
"datecollected" : "1985-03-14",
"recordids" : [
"urn:uuid:5f5e476f-ee39-41c9-8b25-c929f85a773a"
],
"geopoint" : {
"lat" : 23.5821004818154,
"lon" : -105.859665870667
},
"uuid" : "0cec43d8-9ad5-478a-bea1-397ab5cc4430",
"locality" : "ca. 1 mi n of hwy 40 between villa union and cd. durango, and ca. 5 mi w of durango state border.",
"minelevation" : 2316,
"collectionid" : "urn:uuid:a2e32c87-d320-4a01-bafd-a9182ae2e191",
"datecollected_rejected" : false,
"datemodified_rejected" : false,
"recordset" : "40250f4d-7aa6-4fcc-ac38-2868fa4846bd",
"mediarecords" : [
"db38f86b-99f5-4dc4-b4a6-a045a9c35323"
],
"stateprovince" : "sinaloa",
"country" : "mexico",
"collector" : "t. f. daniel;",
"geopoint_rejected" : false,
"hasImage" : true,
"genus" : "abies",
"data" : {
"idigbio:uuid" : "0cec43d8-9ad5-478a-bea1-397ab5cc4430",
"idigbio:etag" : "2682a8f2fb77d59d50442925a80fe58a2ad51e9c",
"idigbio:links" : {
"mediarecord" : [
"http://api.idigbio.org/v1/mediarecords/db38f86b-99f5-4dc4-b4a6-a045a9c35323"
],
"owner" : [
"872733a2-67a3-4c54-aa76-862735a5f334"
],
"recordset" : [
"http://api.idigbio.org/v1/recordsets/40250f4d-7aa6-4fcc-ac38-2868fa4846bd"
]
},
"idigbio:version" : 1,
"idigbio:createdBy" : "872733a2-67a3-4c54-aa76-862735a5f334",
"idigbio:recordIds" : [
"urn:uuid:5f5e476f-ee39-41c9-8b25-c929f85a773a"
],
"idigbio:data" : {
"dwc:catalogNumber" : "ASU0000166",
"dwc:basisOfRecord" : "PreservedSpecimen",
"symbiotaverbatimScientificName" : "Abies",
"idigbio:links" : {
"mediarecord" : [
"http://api.idigbio.org/v1/mediarecords/db38f86b-99f5-4dc4-b4a6-a045a9c35323"
],
"owner" : [
"872733a2-67a3-4c54-aa76-862735a5f334"
],
"recordset" : [
"http://api.idigbio.org/v1/recordsets/40250f4d-7aa6-4fcc-ac38-2868fa4846bd"
]
},
"dwc:collectionCode" : "Plants",
"dwc:decimalLongitude" : "-105.859665870667",
"dcterms:references" : "http://swbiodiversity.org/seinet/collections/individual/index.php?occid=784316",
"dwc:scientificNameAuthorship" : "P. Mill.",
"dwc:collectionID" : "urn:uuid:a2e32c87-d320-4a01-bafd-a9182ae2e191",
"dwc:minimumElevationInMeters" : "2316",
"dwc:georeferenceSources" : "georef batch tool 2013-05-10",
"dwc:georeferenceVerificationStatus" : "reviewed - high confidence",
"dwc:occurrenceRemarks" : "Tree to 4 m; occasional in N-facing barranca",
"dwc:otherCatalogNumbers" : "153418",
"dwc:rights" : "http://creativecommons.org/licenses/by-nc-sa/3.0/",
"idigbio:version" : 1,
"idigbio:recordIds" : [
"urn:uuid:5f5e476f-ee39-41c9-8b25-c929f85a773a"
],
"dwc:decimalLatitude" : "23.5821004818154",
"dwc:day" : "14",
"idigbio:recordId" : "urn:uuid:5f5e476f-ee39-41c9-8b25-c929f85a773a",
"dwc:locality" : "Ca. 1 mi N of Hwy 40 between Villa Union and Cd. Durango, and ca. 5 mi W of Durango state border.",
"dwc:occurrenceID" : "784316",
"idigbio:etag" : "2682a8f2fb77d59d50442925a80fe58a2ad51e9c",
"dwc:year" : "1985",
"dwc:recordedBy" : "T. F. Daniel; ",
"dwc:recordNumber" : "4031",
"dwc:georeferencedBy" : "cjsdavis",
"dwc:scientificName" : "Abies",
"dwc:family" : "Pinaceae",
"dcterms:modified" : "2013-05-10 13:39:12",
"idigbio:createdBy" : "872733a2-67a3-4c54-aa76-862735a5f334",
"dwc:country" : "Mexico",
"idigbio:dateModified" : "2014-04-20T23:11:09.437Z",
"dwc:eventDate" : "1985-03-14",
"dwc:verbatimElevation" : "7600ft",
"idigbio:uuid" : "0cec43d8-9ad5-478a-bea1-397ab5cc4430",
"dwc:startDayOfYear" : "73",
"dwc:habitat" : "Pine-oak forest, in N-facing barranca.",
"dwc:dynamicProperties" : "Tree to 4 m; occasional in N-facing barranca",
"dwc:month" : "3",
"dwc:stateProvince" : "Sinaloa",
"dwc:genus" : "Abies",
"dwc:coordinateUncertaintyInMeters" : "1000",
"dwc:institutionCode" : "ASU"
},
"idigbio:dateModified" : "2014-04-20T23:11:09.437Z"
},
"datemodified" : "2014-04-20",
"scientificname" : "abies",
"minelevation_rejected" : false,
"catalognumber" : "asu0000166",
"etag" : "2682a8f2fb77d59d50442925a80fe58a2ad51e9c",
"mindepth_rejected" : false,
"maxelevation_rejected" : false,
"maxdepth_rejected" : false,
"recordnumber" : "4031",
"family" : "pinaceae",
"institutioncode" : "asu"
},
"_score" : 14.687269,
"_index" : "idigbio-1.1.0",
"_id" : "0cec43d8-9ad5-478a-bea1-397ab5cc4430",
"_type" : "records"
}
],
"max_score" : 14.687269,
"total" : 1
},
"timed_out" : false,
"_shards" : {
"failed" : 0,
"successful" : 10,
"total" : 10
},
"took" : 5
}
Use the Elasticsearch Request Body Search
A JSON query document can be specified on the command-line. Again we are searching for the iDigBio UUID in the "uuid" term.
The following command sends the query as an HTTP "POST" request:
$ curl -s https://search.idigbio.org/idigbio/records/_search -d'
{
"filter" : {
"term" : { "uuid" : "0cec43d8-9ad5-478a-bea1-397ab5cc4430" }
}
}
' | json_pp
and gives the following result:
{
"hits" : {
"hits" : [
{
"_source" : {
"collectioncode" : "plants",
"datecollected" : "1985-03-14",
"recordids" : [
"urn:uuid:5f5e476f-ee39-41c9-8b25-c929f85a773a"
],
"geopoint" : {
"lat" : 23.5821004818154,
"lon" : -105.859665870667
},
"uuid" : "0cec43d8-9ad5-478a-bea1-397ab5cc4430",
"locality" : "ca. 1 mi n of hwy 40 between villa union and cd. durango, and ca. 5 mi w of durango state border.",
"minelevation" : 2316,
"collectionid" : "urn:uuid:a2e32c87-d320-4a01-bafd-a9182ae2e191",
"datecollected_rejected" : false,
"datemodified_rejected" : false,
"recordset" : "40250f4d-7aa6-4fcc-ac38-2868fa4846bd",
"mediarecords" : [
"db38f86b-99f5-4dc4-b4a6-a045a9c35323"
],
"stateprovince" : "sinaloa",
"country" : "mexico",
"collector" : "t. f. daniel;",
"geopoint_rejected" : false,
"hasImage" : true,
"genus" : "abies",
"data" : {
"idigbio:uuid" : "0cec43d8-9ad5-478a-bea1-397ab5cc4430",
"idigbio:etag" : "2682a8f2fb77d59d50442925a80fe58a2ad51e9c",
"idigbio:links" : {
"mediarecord" : [
"http://api.idigbio.org/v1/mediarecords/db38f86b-99f5-4dc4-b4a6-a045a9c35323"
],
"owner" : [
"872733a2-67a3-4c54-aa76-862735a5f334"
],
"recordset" : [
"http://api.idigbio.org/v1/recordsets/40250f4d-7aa6-4fcc-ac38-2868fa4846bd"
]
},
"idigbio:version" : 1,
"idigbio:createdBy" : "872733a2-67a3-4c54-aa76-862735a5f334",
"idigbio:recordIds" : [
"urn:uuid:5f5e476f-ee39-41c9-8b25-c929f85a773a"
],
"idigbio:data" : {
"dwc:catalogNumber" : "ASU0000166",
"dwc:basisOfRecord" : "PreservedSpecimen",
"symbiotaverbatimScientificName" : "Abies",
"idigbio:links" : {
"mediarecord" : [
"http://api.idigbio.org/v1/mediarecords/db38f86b-99f5-4dc4-b4a6-a045a9c35323"
],
"owner" : [
"872733a2-67a3-4c54-aa76-862735a5f334"
],
"recordset" : [
"http://api.idigbio.org/v1/recordsets/40250f4d-7aa6-4fcc-ac38-2868fa4846bd"
]
},
"dwc:collectionCode" : "Plants",
"dwc:decimalLongitude" : "-105.859665870667",
"dcterms:references" : "http://swbiodiversity.org/seinet/collections/individual/index.php?occid=784316",
"dwc:scientificNameAuthorship" : "P. Mill.",
"dwc:collectionID" : "urn:uuid:a2e32c87-d320-4a01-bafd-a9182ae2e191",
"dwc:minimumElevationInMeters" : "2316",
"dwc:georeferenceSources" : "georef batch tool 2013-05-10",
"dwc:georeferenceVerificationStatus" : "reviewed - high confidence",
"dwc:occurrenceRemarks" : "Tree to 4 m; occasional in N-facing barranca",
"dwc:otherCatalogNumbers" : "153418",
"dwc:rights" : "http://creativecommons.org/licenses/by-nc-sa/3.0/",
"idigbio:version" : 1,
"idigbio:recordIds" : [
"urn:uuid:5f5e476f-ee39-41c9-8b25-c929f85a773a"
],
"dwc:decimalLatitude" : "23.5821004818154",
"dwc:day" : "14",
"idigbio:recordId" : "urn:uuid:5f5e476f-ee39-41c9-8b25-c929f85a773a",
"dwc:locality" : "Ca. 1 mi N of Hwy 40 between Villa Union and Cd. Durango, and ca. 5 mi W of Durango state border.",
"dwc:occurrenceID" : "784316",
"idigbio:etag" : "2682a8f2fb77d59d50442925a80fe58a2ad51e9c",
"dwc:year" : "1985",
"dwc:recordedBy" : "T. F. Daniel; ",
"dwc:recordNumber" : "4031",
"dwc:georeferencedBy" : "cjsdavis",
"dwc:scientificName" : "Abies",
"dwc:family" : "Pinaceae",
"dcterms:modified" : "2013-05-10 13:39:12",
"idigbio:createdBy" : "872733a2-67a3-4c54-aa76-862735a5f334",
"dwc:country" : "Mexico",
"idigbio:dateModified" : "2014-04-20T23:11:09.437Z",
"dwc:eventDate" : "1985-03-14",
"dwc:verbatimElevation" : "7600ft",
"idigbio:uuid" : "0cec43d8-9ad5-478a-bea1-397ab5cc4430",
"dwc:startDayOfYear" : "73",
"dwc:habitat" : "Pine-oak forest, in N-facing barranca.",
"dwc:dynamicProperties" : "Tree to 4 m; occasional in N-facing barranca",
"dwc:month" : "3",
"dwc:stateProvince" : "Sinaloa",
"dwc:genus" : "Abies",
"dwc:coordinateUncertaintyInMeters" : "1000",
"dwc:institutionCode" : "ASU"
},
"idigbio:dateModified" : "2014-04-20T23:11:09.437Z"
},
"datemodified" : "2014-04-20",
"scientificname" : "abies",
"minelevation_rejected" : false,
"catalognumber" : "asu0000166",
"etag" : "2682a8f2fb77d59d50442925a80fe58a2ad51e9c",
"mindepth_rejected" : false,
"maxelevation_rejected" : false,
"maxdepth_rejected" : false,
"recordnumber" : "4031",
"family" : "pinaceae",
"institutioncode" : "asu"
},
"_score" : 1,
"_index" : "idigbio-1.1.0",
"_id" : "0cec43d8-9ad5-478a-bea1-397ab5cc4430",
"_type" : "records"
}
],
"max_score" : 1,
"total" : 1
},
"timed_out" : false,
"_shards" : {
"failed" : 0,
"successful" : 10,
"total" : 10
},
"took" : 37
}
Alternatively, the JSON-formatted query body may be sourced from a file. In this example, the file is named uuid.json and contains the iDigBio UUID in the "uuid" term as before:
$ cat uuid.json
{
"filter" : {
"term" : { "uuid" : "0cec43d8-9ad5-478a-bea1-397ab5cc4430" }
}
}
We tell curl to use the file as the source of the data with the at '@' sign.
$ curl -s -XGET https://search.idigbio.org/idigbio/records/_search -d@uuid.json | json_pp
{
"hits" : {
"hits" : [
{
"_source" : {
"collectioncode" : "plants",
"datecollected" : "1985-03-14",
"uuid" : "0cec43d8-9ad5-478a-bea1-397ab5cc4430",
"geopoint_geohash_6" : "9shwph",
"locality" : "ca. 1 mi n of hwy 40 between villa union and cd. durango, and ca. 5 mi w of durango state border.",
"minelevation" : 231,
"collectionid" : "urn:uuid:a2e32c87-d320-4a01-bafd-a9182ae2e191",
"datecollected_rejected" : false,
"datemodified_rejected" : false,
"geopoint_geohash_5" : "9shwp",
"recordset" : "40250f4d-7aa6-4fcc-ac38-2868fa4846bd",
"geopoint_rejected" : false,
"hasImage" : true,
"data" : {
"idigbio:uuid" : "0cec43d8-9ad5-478a-bea1-397ab5cc4430",
"idigbio:etag" : "2682a8f2fb77d59d50442925a80fe58a2ad51e9c",
"idigbio:links" : {
"mediarecord" : [
"http://api.idigbio.org/v1/mediarecords/db38f86b-99f5-4dc4-b4a6-a045a9c35323"
],
"owner" : [
"872733a2-67a3-4c54-aa76-862735a5f334"
],
"recordset" : [
"http://api.idigbio.org/v1/recordsets/40250f4d-7aa6-4fcc-ac38-2868fa4846bd"
]
},
"idigbio:version" : 1,
"idigbio:createdBy" : "872733a2-67a3-4c54-aa76-862735a5f334",
"idigbio:recordIds" : [
"urn:uuid:5f5e476f-ee39-41c9-8b25-c929f85a773a"
],
"idigbio:data" : {
"dwc:catalogNumber" : "ASU0000166",
"dwc:basisOfRecord" : "PreservedSpecimen",
"symbiotaverbatimScientificName" : "Abies",
"idigbio:links" : {
"mediarecord" : [
"http://api.idigbio.org/v1/mediarecords/db38f86b-99f5-4dc4-b4a6-a045a9c35323"
],
"owner" : [
"872733a2-67a3-4c54-aa76-862735a5f334"
],
"recordset" : [
"http://api.idigbio.org/v1/recordsets/40250f4d-7aa6-4fcc-ac38-2868fa4846bd"
]
},
"dwc:collectionCode" : "Plants",
"dwc:decimalLongitude" : "-105.859665870667",
"dcterms:references" : "http://swbiodiversity.org/seinet/collections/individual/index.php?occid=784316",
"dwc:scientificNameAuthorship" : "P. Mill.",
"dwc:collectionID" : "urn:uuid:a2e32c87-d320-4a01-bafd-a9182ae2e191",
"dwc:minimumElevationInMeters" : "2316",
"dwc:georeferenceSources" : "georef batch tool 2013-05-10",
"dwc:georeferenceVerificationStatus" : "reviewed - high confidence",
"dwc:occurrenceRemarks" : "Tree to 4 m; occasional in N-facing barranca",
"dwc:otherCatalogNumbers" : "153418",
"dwc:rights" : "http://creativecommons.org/licenses/by-nc-sa/3.0/",
"idigbio:version" : 1,
"idigbio:recordIds" : [
"urn:uuid:5f5e476f-ee39-41c9-8b25-c929f85a773a"
],
"dwc:decimalLatitude" : "23.5821004818154",
"dwc:day" : "14",
"idigbio:recordId" : "urn:uuid:5f5e476f-ee39-41c9-8b25-c929f85a773a",
"dwc:locality" : "Ca. 1 mi N of Hwy 40 between Villa Union and Cd. Durango, and ca. 5 mi W of Durango state border.",
"dwc:occurrenceID" : "784316",
"idigbio:etag" : "2682a8f2fb77d59d50442925a80fe58a2ad51e9c",
"dwc:year" : "1985",
"dwc:recordedBy" : "T. F. Daniel; ",
"dwc:recordNumber" : "4031",
"dwc:georeferencedBy" : "cjsdavis",
"dwc:scientificName" : "Abies",
"dwc:family" : "Pinaceae",
"dcterms:modified" : "2013-05-10 13:39:12",
"idigbio:createdBy" : "872733a2-67a3-4c54-aa76-862735a5f334",
"dwc:country" : "Mexico",
"idigbio:dateModified" : "2014-04-20T23:11:09.437Z",
"dwc:eventDate" : "1985-03-14",
"dwc:verbatimElevation" : "7600ft",
"idigbio:uuid" : "0cec43d8-9ad5-478a-bea1-397ab5cc4430",
"dwc:startDayOfYear" : "73",
"dwc:habitat" : "Pine-oak forest, in N-facing barranca.",
"dwc:dynamicProperties" : "Tree to 4 m; occasional in N-facing barranca",
"dwc:month" : "3",
"dwc:stateProvince" : "Sinaloa",
"dwc:genus" : "Abies",
"dwc:coordinateUncertaintyInMeters" : "1000",
"dwc:institutionCode" : "ASU"
},
"idigbio:dateModified" : "2014-04-20T23:11:09.437Z"
},
"scientificname" : "abies",
"minelevation_rejected" : false,
"etag" : "2682a8f2fb77d59d50442925a80fe58a2ad51e9c",
"maxdepth_rejected" : false,
"family" : "pinaceae",
"institutioncode" : "asu",
"geopoint" : {
"lat" : 23.5821004818154,
"lon" : -105.859665870667
},
"fieldnumber" : "4031",
"geopoint_geohash_2" : "9s",
"stateprovince" : "sinaloa",
"mediarecords" : [
"db38f86b-99f5-4dc4-b4a6-a045a9c35323"
],
"country" : "mexico",
"collector" : "t. f. daniel;",
"geopoint_geohash_4" : "9shw",
"version" : 1,
"genus" : "abies",
"geopoint_geohash_3" : "9sh",
"datemodified" : "2014-04-20",
"catalognumber" : "asu0000166",
"mindepth_rejected" : false,
"maxelevation_rejected" : false
},
"_score" : 1,
"_index" : "idigbio-1.0.0",
"_id" : "0cec43d8-9ad5-478a-bea1-397ab5cc4430",
"_type" : "records"
}
],
"max_score" : 1,
"total" : 1
},
"timed_out" : false,
"_shards" : {
"failed" : 0,
"successful" : 10,
"total" : 10
},
"took" : 64
}
Fetch an Older Version of a Particular Record
Consider an iDigBio Media Record that is identified by:
"idigbio:uuid" : "ff5fd841-6bc6-4241-80d3-def7e1b6d5e8"
This record has been updated over time so there are multiple revisions of the record preserved in the API. We can find that the most recent (e.g. "current") version of this particular record is version 1:
$ curl -s http://api.idigbio.org/v1/mediarecords/ff5fd841-6bc6-4241-80d3-def7e1b6d5e8 | json_pp | grep version "idigbio:version" : 1,
We would like to look at the previous version (version 0) to see what has changed. We do this by adding the "version" parameter to the url:
curl -s http://api.idigbio.org/v1/mediarecords/ff5fd841-6bc6-4241-80d3-def7e1b6d5e8?version=0 | json_pp | grep version "idigbio:version" : "0",
The "version" parameter is available for any most of the endpoints that return a single entity record (as opposed to a List).
Paging through the API
Endpoints that return a list of entities may have more total items than are able to be "fit" in a single request. For example, trying to fetch the list of all records in iDigBio will surely timeout in the browser before it is completed (and would put undue load on the server infrastructure).
Request 5 media record entities:
$ curl -s "http://api.idigbio.org/v1/mediarecords?limit=5" | json_pp
{
"idigbio:errors" : [],
"idigbio:links" : {
"idigbio:nextPage" : "http://api.idigbio.org/v1/mediarecords?limit=5&offset=5"
},
"idigbio:items" : [
{
"idigbio:links" : {
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/000003cd-0cca-421b-8f26-f557a26b0393"
},
"idigbio:uuid" : "000003cd-0cca-421b-8f26-f557a26b0393",
"idigbio:version" : 1,
"idigbio:etag" : "ce3e2f7272ec996bb479c87549ba90c15ba96426",
"idigbio:dateModified" : "2014-04-21T22:19:27.436Z"
},
{
"idigbio:links" : {
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/00000728-ffb3-4a68-9f93-137f19961121"
},
"idigbio:uuid" : "00000728-ffb3-4a68-9f93-137f19961121",
"idigbio:version" : 3,
"idigbio:etag" : "ef2cac326a60d89d8cb9005abaa82068bfa83565",
"idigbio:dateModified" : "2014-04-24T05:03:56.782Z"
},
{
"idigbio:links" : {
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/00000b03-e208-4d22-983b-506ad2842f7c"
},
"idigbio:uuid" : "00000b03-e208-4d22-983b-506ad2842f7c",
"idigbio:version" : 2,
"idigbio:etag" : "bc118a7ea53e004c82ab9b7e813e1010ae5f8e17",
"idigbio:dateModified" : "2014-04-20T05:16:20.389Z"
},
{
"idigbio:links" : {
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/000010bc-a4d4-483d-b71d-0dbdd4fd2d5a"
},
"idigbio:uuid" : "000010bc-a4d4-483d-b71d-0dbdd4fd2d5a",
"idigbio:version" : 0,
"idigbio:etag" : "68c441bd3c49507bf930f3b278f2c58f9cb792ec",
"idigbio:dateModified" : "2014-04-20T21:38:46.679Z"
},
{
"idigbio:links" : {
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/000012f9-d288-4a14-b898-77430e0a137a"
},
"idigbio:uuid" : "000012f9-d288-4a14-b898-77430e0a137a",
"idigbio:version" : 1,
"idigbio:etag" : "cf49416750fdb9bdb808c334a74b84f27bb8160b",
"idigbio:dateModified" : "2014-04-23T02:43:08.344Z"
}
],
"idigbio:itemCount" : "2342880"
}
The next "page" of records can be requested by adding the "offset" paramenter.
A link that includes the offset to the next page is provided in the results:
"idigbio:links" : {
"idigbio:nextPage" : "http://api.idigbio.org/v1/mediarecords?limit=5&offset=5"
}
Request the next "page" of entity ids by starting at offset 5.
$ curl -s "http://api.idigbio.org/v1/mediarecords?limit=5&offset=5" | json_pp
{
"idigbio:errors" : [],
"idigbio:links" : {
"idigbio:nextPage" : "http://api.idigbio.org/v1/mediarecords?limit=5&offset=10",
"idigbio:prevPage" : "http://api.idigbio.org/v1/mediarecords?limit=5&offset=0"
},
"idigbio:items" : [
{
"idigbio:links" : {
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/00001478-c150-4faf-a617-439a838d4377"
},
"idigbio:uuid" : "00001478-c150-4faf-a617-439a838d4377",
"idigbio:version" : 1,
"idigbio:etag" : "30f602e4eb47ebb2ceb265f64217e3cf5664f517",
"idigbio:dateModified" : "2014-03-21T23:09:39.752Z"
},
{
"idigbio:links" : {
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/00001a91-189b-4002-b56e-a770a55951a0"
},
"idigbio:uuid" : "00001a91-189b-4002-b56e-a770a55951a0",
"idigbio:version" : 0,
"idigbio:etag" : "647e82d17ee435fb14f0f8607dabe88dfc3a1944",
"idigbio:dateModified" : "2014-04-25T04:49:32.359Z"
},
{
"idigbio:links" : {
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/00002091-4fb3-410a-9307-bd3e917dfcca"
},
"idigbio:uuid" : "00002091-4fb3-410a-9307-bd3e917dfcca",
"idigbio:version" : 0,
"idigbio:etag" : "90d98d48d9e7e07eab9064bd9b6e22ce6502c07f",
"idigbio:dateModified" : "2014-05-03T18:45:47.112Z"
},
{
"idigbio:links" : {
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/00002c32-ae3a-41ed-9bd9-f6c50d3e35fb"
},
"idigbio:uuid" : "00002c32-ae3a-41ed-9bd9-f6c50d3e35fb",
"idigbio:version" : 3,
"idigbio:etag" : "d1ded90d06e93876b1badd01222905add93e8806",
"idigbio:dateModified" : "2014-04-19T00:25:59.471Z"
},
{
"idigbio:links" : {
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/00002dbd-6415-463b-8cae-38f548415ffa"
},
"idigbio:uuid" : "00002dbd-6415-463b-8cae-38f548415ffa",
"idigbio:version" : 2,
"idigbio:etag" : "4e298045b496146f5c51e331c9887fd7afde4deb",
"idigbio:dateModified" : "2014-04-21T20:29:39.531Z"
}
],
"idigbio:itemCount" : "2342880"
}
which includes links to the previous page and next page:
"idigbio:nextPage" : "http://api.idigbio.org/v1/mediarecords?limit=5&offset=10",
"idigbio:prevPage" : "http://api.idigbio.org/v1/mediarecords?limit=5&offset=0"
With current results starting at offset 5 and a limit of 5, the previous page and a next page offsets are 0 and 10 respectively (offset +- limit).
- TO DO One more paging to help differentiate these multiples of 5.
Fetch a list of five MediaRecord IDs
The "mediarecords" endpoint returns a List of Media Record IDs.
Example URL:
http://api.idigbio.org/v1/mediarecords?limit=5&offset=5
Example Results:
$ curl -s "http://api.idigbio.org/v1/mediarecords?limit=5&offset=5" | json_pp
{
"idigbio:errors" : [],
"idigbio:links" : {
"idigbio:nextPage" : "http://api.idigbio.org/v1/mediarecords?limit=5&offset=10",
"idigbio:prevPage" : "http://api.idigbio.org/v1/mediarecords?limit=5&offset=0"
},
"idigbio:items" : [
{
"idigbio:links" : {
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/00001478-c150-4faf-a617-439a838d4377"
},
"idigbio:uuid" : "00001478-c150-4faf-a617-439a838d4377",
"idigbio:version" : 1,
"idigbio:etag" : "30f602e4eb47ebb2ceb265f64217e3cf5664f517",
"idigbio:dateModified" : "2014-03-21T23:09:39.752Z"
},
{
"idigbio:links" : {
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/00001a91-189b-4002-b56e-a770a55951a0"
},
"idigbio:uuid" : "00001a91-189b-4002-b56e-a770a55951a0",
"idigbio:version" : 0,
"idigbio:etag" : "647e82d17ee435fb14f0f8607dabe88dfc3a1944",
"idigbio:dateModified" : "2014-04-25T04:49:32.359Z"
},
{
"idigbio:links" : {
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/00002091-4fb3-410a-9307-bd3e917dfcca"
},
"idigbio:uuid" : "00002091-4fb3-410a-9307-bd3e917dfcca",
"idigbio:version" : 0,
"idigbio:etag" : "90d98d48d9e7e07eab9064bd9b6e22ce6502c07f",
"idigbio:dateModified" : "2014-05-03T18:45:47.112Z"
},
{
"idigbio:links" : {
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/00002c32-ae3a-41ed-9bd9-f6c50d3e35fb"
},
"idigbio:uuid" : "00002c32-ae3a-41ed-9bd9-f6c50d3e35fb",
"idigbio:version" : 3,
"idigbio:etag" : "d1ded90d06e93876b1badd01222905add93e8806",
"idigbio:dateModified" : "2014-04-19T00:25:59.471Z"
},
{
"idigbio:links" : {
"mediarecord" : "http://api.idigbio.org/v1/mediarecords/00002dbd-6415-463b-8cae-38f548415ffa"
},
"idigbio:uuid" : "00002dbd-6415-463b-8cae-38f548415ffa",
"idigbio:version" : 2,
"idigbio:etag" : "4e298045b496146f5c51e331c9887fd7afde4deb",
"idigbio:dateModified" : "2014-04-21T20:29:39.531Z"
}
],
"idigbio:itemCount" : "2342880"
}
Screenshot of Results:
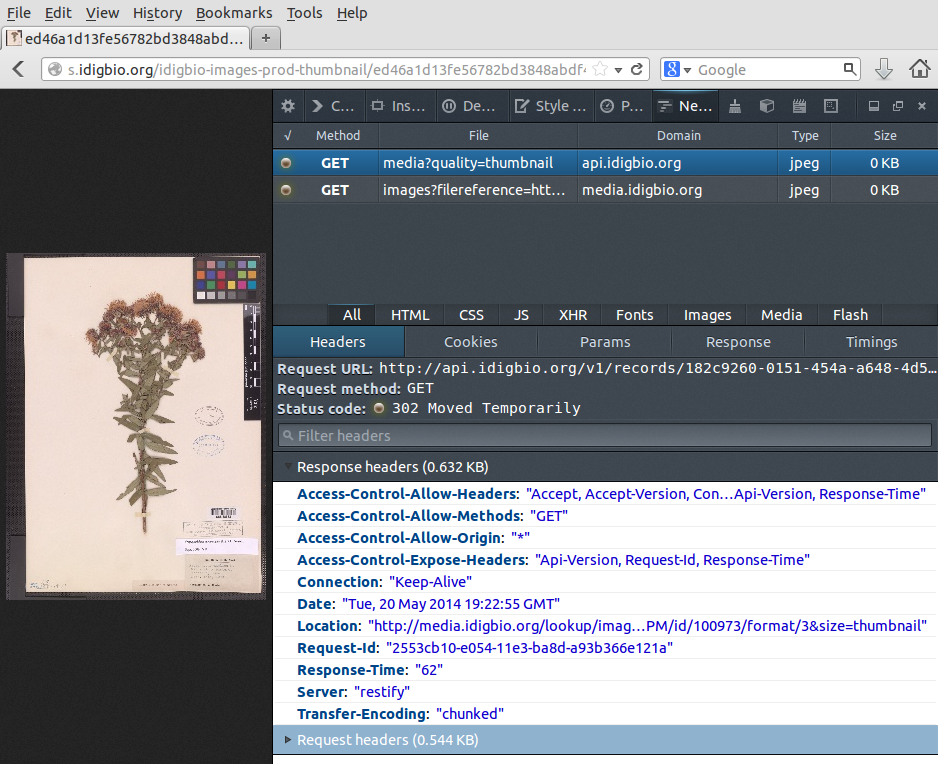
Display a thumbnail image for a specimen record
Let's say that we wish to display a thumbnail image for a particular specimen record.
We can look at the full record:
$ curl -s "http://api.idigbio.org/v1/records/182c9260-0151-454a-a648-4d58ddf51bd8" | json_pp
{
"idigbio:uuid" : "182c9260-0151-454a-a648-4d58ddf51bd8",
"idigbio:etag" : "bb5ee46e2730948c946c178e8da85dedee4bea80",
"idigbio:links" : {
"mediarecord" : [
"http://api.idigbio.org/v1/mediarecords/00002091-4fb3-410a-9307-bd3e917dfcca"
],
"owner" : [
"872733a2-67a3-4c54-aa76-862735a5f334"
],
"recordset" : [
"http://api.idigbio.org/v1/recordsets/09edf7d2-e68e-4a42-93da-762f86bb814f"
]
},
"idigbio:version" : 0,
"idigbio:createdBy" : "872733a2-67a3-4c54-aa76-862735a5f334",
"idigbio:recordIds" : [
"urn:uuid:7d0d4400-7c6c-4bb4-b2ad-7b1f6e776755"
],
"idigbio:dateModified" : "2014-05-03T14:27:40.596Z",
"idigbio:data" : {
"idigbio:recordId" : "urn:uuid:7d0d4400-7c6c-4bb4-b2ad-7b1f6e776755",
"dwc:catalogNumber" : "CBS.038533",
"dwc:specificEpithet" : "novae-angliae",
"dwc:occurrenceID" : "272024",
"dwc:year" : "0",
"dwc:recordedBy" : "Hugh S. Clark",
"dwc:scientificName" : "Symphyotrichum novae-angliae",
"dwc:basisOfRecord" : "PreservedSpecimen",
"dwc:family" : "Asteraceae",
"symbiotaverbatimScientificName" : "Symphyotrichum novae-angliae",
"dwc:collectionCode" : "CBS",
"dcterms:modified" : "2012-03-14 00:00:00",
"dwc:decimalLongitude" : "-72.6834",
"dwc:country" : "USA",
"dcterms:references" : "http://portal.neherbaria.org/portal/collections/individual/index.php?occid=272024",
"dwc:municipality" : "Hartford",
"dwc:scientificNameAuthorship" : "(L.) G.L. Nesom",
"dwc:month" : "0",
"dwc:rights" : "http://hdl.handle.net/10079/8931zqj",
"dwc:geodeticDatum" : "WGS84",
"dwc:rightsHolder" : "Peabody Museum of Natural History, Yale University",
"dwc:stateProvince" : "Connecticut",
"dwc:genus" : "Symphyotrichum",
"dwc:coordinateUncertaintyInMeters" : "5419",
"dwc:decimalLatitude" : "41.766",
"dwc:accessRights" : "http://hdl.handle.net/10079/8931zqj",
"dwc:institutionCode" : "YPM",
"dwc:county" : "Hartford County"
}
}
At this point we have a couple of options to get to the thumbnail. The easiest is to use the /media endpoint for this record's iDigBio UUID and add the "quality" parameter with "thumbnail" as the value:
http://api.idigbio.org/v1/records/182c9260-0151-454a-a648-4d58ddf51bd8/media?quality=thumbnail
which results in a number of HTTP Redirects to the target thumbnail image, illustrated here with the Chrome Developer Tools visible:
Note that the final image URL should not be considered stable or permanent. Images can be moved around to various storage platforms. The Mediarecord has a UUID that will remain contstant but the containing data, including the image location, may be updated over time.
The /media endpoint of a specimen record is kind of shortcut to avoid having to walk through the relationship to a mediarecord and then to an image file.
Alternatively, if we actually wanted to look at media metadata, we could use the link to the mediarecord in the "idigbio:links" section:
"idigbio:links" : {
"mediarecord" : [
"http://api.idigbio.org/v1/mediarecords/00002091-4fb3-410a-9307-bd3e917dfcca"
],
which also has a /media endpoint that can lead to the same thumbnail image.
http://api.idigbio.org/v1/mediarecords/00002091-4fb3-410a-9307-bd3e917dfcca/media?quality=thumbnail
Advanced Elasticsearch Examples
| Direct queries to the iDigBio Elasticsearch service are deprecated. The iDigBio API v2 / iDigBio Search API specification should be used instead. |
Search using Locality Data
The iDigBio Elasticsearch documentation should be considered required reading for these advanced Elasticsearch examples.
- 1. Find all specimen records that meet the following criteria
- Has an image (mediarecord) associated with the specimen record
- Is from Hocking County, Ohio or Fairfield County, Ohio.
Note that we are not using georeference data here, we are using the "county" and "stateprovince" fields.
To resolve this criteria, we need the following (expressed in pseudocode):
- "hasImage" is true
- "county" is either "Hocking" or "Fairfield"
- "stateprovince" is "Ohio" (there are multiple Fairfield counties throughout the world so we need to restrict the geography by state)
The essential query parameter string will look something like this (note we must lowercase the values):
?q=hasImage:true+AND+county:(fairfield+OR+hocking)+AND+stateprovince:ohio
In addition, it is possible to request more or fewer records at a time using the "from" and "size" parameters. Below is the full curl example requesting only the first 3 records and using grep on the output to filter everything except the iDigBio UUID field. Note that we quote the full URL to keep the shell from trying to interpret the special characters.
$ curl -s 'http://search.idigbio.org/idigbio/records/_search?from=0&size=3&q=hasImage:true+AND+county:(fairfield+OR+hocking)+AND+stateprovince:ohio' | json_pp | egrep "\"uuid\""
"uuid" : "ecf14f53-d225-4eb5-89e6-66379b360781",
"uuid" : "e45071cd-40eb-41e7-9105-0e35234025f5",
"uuid" : "20ad60a8-4126-4cc0-9378-411a9a640005",
Now that we have iDigBio UUIDs, we could retrieve images for these specimens by iterating over the /media endpoints for each of those uuids in the API.
http://api.idigbio.org/v1/records/ecf14f53-d225-4eb5-89e6-66379b360781/media?quality=webview
http://api.idigbio.org/v1/records/e45071cd-40eb-41e7-9105-0e35234025f5/media?quality=webview
...
Note that we did not specify any sorting in the Elasticsearch query and we did restrict the result count by using the "size" parameter. If the same query is run at a later time the specific records returned could be different.
Note also that the above query is for an exact match on the county name which will perform fairly well. Unfortunately, the data includes many variations in the county field for these counties such as:
"Fairfield" "Fairfield County" "Hocking" "Hocking Co." "Hocking County" "Hocking Hills"
To find these related records, a wildcard query may be useful. In the following case the size is also increased to 3000 to retrieve a larger sample.
$ curl -s 'http://search.idigbio.org/idigbio/records/_search?from=0&size=3000&q=hasImage:true+AND+county:(fairfield*+OR+hocking*)+AND+stateprovince:ohio' | json_pp
Family: Curculionidae - Search Example
| Direct queries to the iDigBio Elasticsearch service are deprecated. The iDigBio API v2 / iDigBio Search API specification should be used instead. |
Let us say we are looking for images associated with specimen records for Family Curculionidae (a family of weevils).
If we already know the iDigBio UUID of a specimen we can simply use the /media endpoint for the record:
http://api.idigbio.org/v1/records/c2e34bda-0405-48ea-b7b2-76f893777f20/media?quality=webview
Giving us an image from the iDigBio API that looks something like this:
But how did we find the proper iDigBio UUID for this specimen record in the first place?
The iDigBio Portal Search is of course built just for this purpose.
If on the other hand we wanted to search for these specimens via a software program, we could use the iDigBio Elasticsearch interface.
I happen to know that this specimen was collected in New Jersey, so we search for records of Family Curculionidae in New Jersey that have associated images. Note the special encoding of the query string (space becomes %20, quotation becomes %22) required when using curl vs. a browser (the browser would do this encoding for you).
The following example may not be correct yet...
$ curl -s 'http://search.idigbio.org/idigbio/records/_search?q=hasImage:true+AND+family:curculionidae+AND+stateprovince:%22new%20jersey%22'
Mapping Example
| Direct queries to the iDigBio Elasticsearch service are deprecated. The iDigBio API v2 / iDigBio Search API specification should be used instead. |
This Mapping example is really just another Elasticsearch example. To generate a map, we basically want to do an Elasticsearch query to choose our specimens of interest and then put the points for those specimens on a map.
The iDigBio Elasticsearch documentation should be considered required reading before attempting to integrate this code into one's site.
This example displays a map that includes specimen data points that match the criteria of Scientific Name (dwc:scientificName) is "Abietinella abietina" and Institution Code (dwc:institutionCode) is "DUKE".
The Elasticsearch terms for this query are:
scientificname" : "abietinella abietina" institutioncode": "duke"
The following is a full HTML page with javascript code that displays a small specimen map containing iDigBio data points.
To customize the map search results, modify the var terms = [ ... section with the terms of your choice.
<!DOCTYPE html>
<html lang="en">
<head>
<title>iDigBio Map</title>
<link rel="stylesheet" href="http://cdn.leafletjs.com/leaflet-0.7.2/leaflet.css" />
<script src="http://code.jquery.com/jquery-1.11.0.min.js"></script>
<script src="http://cdn.leafletjs.com/leaflet-0.7.2/leaflet.js"></script>
<script type="text/javascript">
/*
* This example provided by iDigBio. www.idigbio.org
*
* The list of available terms is available at
* https://www.idigbio.org/wiki/index.php/IDigBio_API_v1_Specification#Search
*
* Some Example Terms:
* uuid, scientificname, institutioncode, collectioncode, genus,
* country, stateprovince, county, occurenceid, barcodevalue, catalognumber
*****/
var terms = [
{"term" : { "scientificname" : "abietinella abietina"}},
{"term" : { "institutioncode": "duke"}}
];
$(document).ready(function(){
var query = {
"size": 10000,
"from": 0,
"query":{
"filtered":{
"filter":{
"and":[]
}
}
},
"sort":{
"scientificname":{"order":"asc"}
}
};
/*
*Build query params and outlink
***/
var link=[];
$.each(terms,function(ind,item){
query.query.filtered.filter.and.push(item);
$.each(item.term,function(key,val){
link.push(key+'='+ val.toLowerCase());
});
});
var link_href = 'https://www.idigbio.org/portal/search?'+link.join('&');
var link_string = '<br>Map point data provided by iDigBio. <a id="link" href="' + link_href + '">View on iDigBio</a>';
var base = L.tileLayer('http://{s}.tile.openstreetmap.org/{z}/{x}/{y}.png',{
attribution: 'Map data © OpenStreetMap contributors; ' + link_string,
minZoom: 0,
maxZoom: 18,
reuseTiles: true
});
map = L.map('map',{
center: [0,0],
zoom: 0,
layers: [base],
scrollWheelZoom: true,
boxZoom: false
});
/*
*Make request to iDigBio search server
***/
$.post('https://search.idigbio.org/idigbio/records/_search', JSON.stringify(query),function(resp){
$(resp.hits.hits).each(function(ind,item){
var source = item._source;
if(typeof source.geopoint !== 'undefined'){
var c = new L.Circle([source.geopoint.lat,source.geopoint.lon],1,{color:'#E90A0A',opacity:1,fillopacity:1});
c.addTo(map);
}
});
});
});
</script>
</head>
<body>
<div id="map" style="height:250px;width:330px;"></div>
</body>
</html>
The above html page would render as follows (captured in a screenshot) in a browser:
Deeper integration of iDigBio mapping requires understanding of geohashes and would be a more significant undertaking. If your project wishes advanced integration, please contact iDigBio. https://www.idigbio.org/contact
idigbio:links and the 20 Item List
Compare the idigbio:links section of the following two Publisher records:
http://api.idigbio.org/v1/publishers/076c0ff6-65e9-48a5-8e4b-2447936f9a1c
http://api.idigbio.org/v1/publishers/4e1beef9-d7c0-4ac0-87df-065bc5a55361
The first Publisher record returns a JSON document containing "idigbio:links" and a List of recordsets (or to be more precise, a List of API urls that can be used to access individual Recordset records by iDigBio UUID). Since the number of recordsets is less than 20, they are included in the Publisher record output.
$ curl -s http://api.idigbio.org/v1/publishers/076c0ff6-65e9-48a5-8e4b-2447936f9a1c | json_pp
{
"idigbio:uuid" : "076c0ff6-65e9-48a5-8e4b-2447936f9a1c",
"idigbio:etag" : "31b115d5a307027f757554441c93d8e5b8ac8fab",
"idigbio:links" : {
"recordset" : [
"http://api.idigbio.org/v1/recordsets/197f7bb8-213d-4fae-b536-652dd91e56dc",
"http://api.idigbio.org/v1/recordsets/31c140bc-e6f1-4acc-beaf-b825cf288ad9",
"http://api.idigbio.org/v1/recordsets/5aac25d2-bcfb-4084-a700-584311ea539d",
"http://api.idigbio.org/v1/recordsets/5e893602-84ca-4c8c-bac1-99111c777582",
"http://api.idigbio.org/v1/recordsets/6539877e-82dc-485c-ad3d-038f383d5431",
"http://api.idigbio.org/v1/recordsets/69037495-438d-4dba-bf0f-4878073766f1",
"http://api.idigbio.org/v1/recordsets/7fcdca8e-7469-480c-8516-cce4e24c37c9",
"http://api.idigbio.org/v1/recordsets/82541f90-fe8e-4d66-84d8-4fe515dc5533",
"http://api.idigbio.org/v1/recordsets/89eb1ad0-ae60-4e8a-bf34-a53d0423bc80",
"http://api.idigbio.org/v1/recordsets/92dd8c8e-c048-4f0a-9b5d-2ee627d2f553",
"http://api.idigbio.org/v1/recordsets/9ace4c05-d930-45c7-8d2d-0cadff1ea32b",
"http://api.idigbio.org/v1/recordsets/9bd4ff72-1cb2-431f-bf7b-b5d47e08cc02",
"http://api.idigbio.org/v1/recordsets/a3b77120-3770-46dd-ba47-6941eff848b3",
"http://api.idigbio.org/v1/recordsets/ab4b6a2b-a90a-44ce-95a1-2c44c911fcc6",
"http://api.idigbio.org/v1/recordsets/cb65cf5e-07b8-4d53-a91a-7dce9b8ccf80",
"http://api.idigbio.org/v1/recordsets/d315c4a3-0bee-49d1-8d03-726358937cde",
"http://api.idigbio.org/v1/recordsets/d767f759-af64-4464-8614-c77ca44cad8d",
"http://api.idigbio.org/v1/recordsets/f83517ea-7b9f-443d-9371-d11a05ebc0a7",
"http://api.idigbio.org/v1/recordsets/ff111763-e72d-4f24-8914-b5b2dd94908c"
]
},
...
The second link returns a JSON document containing "idigbio:links" and a single recordset link rather than a List. To get to the List of recordsets for this Publisher record, this intermediate link needs to be followed.
$ curl -s http://api.idigbio.org/v1/publishers/4e1beef9-d7c0-4ac0-87df-065bc5a55361 | json_pp
{
"idigbio:uuid" : "4e1beef9-d7c0-4ac0-87df-065bc5a55361",
"idigbio:etag" : "01c8eabbf9e8fe7f5af86349b04cad55b3ddee8e",
"idigbio:links" : {
"recordset" : [
"http://api.idigbio.org/v1/publishers/4e1beef9-d7c0-4ac0-87df-065bc5a55361/recordsets"
]
},
...
As suspected, the list of Recordsets includes more than 20 items (in this case, 51 recordset items) which is why they were not displayed with the Publisher record.
$ curl -s http://api.idigbio.org/v1/publishers/4e1beef9-d7c0-4ac0-87df-065bc5a55361/recordsets | json_pp | grep recordset | wc -l
51
$ curl -s http://api.idigbio.org/v1/publishers/4e1beef9-d7c0-4ac0-87df-065bc5a55361/recordsets | json_pp
{
"idigbio:errors" : [],
"idigbio:links" : {},
"idigbio:items" : [
{
"idigbio:links" : {
"recordset" : "http://api.idigbio.org/v1/recordsets/0072bf11-a354-4998-8730-c0cb4cfc9517"
},
"idigbio:uuid" : "0072bf11-a354-4998-8730-c0cb4cfc9517"
},
{
"idigbio:links" : {
"recordset" : "http://api.idigbio.org/v1/recordsets/00d9fcc1-c8e2-4ef6-be64-9994ca6a32c3"
},
"idigbio:uuid" : "00d9fcc1-c8e2-4ef6-be64-9994ca6a32c3"
},
{
"idigbio:links" : {
"recordset" : "http://api.idigbio.org/v1/recordsets/0dd7c8dd-412c-4a13-a1d3-47e1e1af5455"
},
"idigbio:uuid" : "0dd7c8dd-412c-4a13-a1d3-47e1e1af5455"
},
{
"idigbio:links" : {
"recordset" : "http://api.idigbio.org/v1/recordsets/181352ea-3598-4f32-b919-c8f6097f4c65"
},
"idigbio:uuid" : "181352ea-3598-4f32-b919-c8f6097f4c65"
},
{
"idigbio:links" : {
"recordset" : "http://api.idigbio.org/v1/recordsets/196bb137-2f82-40d5-b294-e730af29749f"
},
"idigbio:uuid" : "196bb137-2f82-40d5-b294-e730af29749f"
},
...
API Performance
For large numbers of records or bulk data aquisition, please use the Download features available in the Portal. https://www.idigbio.org/portal
See also iDigBio API Performance.