Data Ingestion Guidance: Difference between revisions
| Line 76: | Line 76: | ||
==Packaging for images/media objects== | ==Packaging for images/media objects== | ||
*Each media record should have a unique (within the dataset) identifier in the ''dwc:occurrenceID'' field. | *Each media record should have a unique (within the dataset) identifier in the ''dwc:occurrenceID'' field. | ||
**If submitting media records with specimen data records, put the specimen ''dwc:occurrenceID'' in the ''ac:associatedSpecimenReference'' field in the image metadata | **If submitting media records with specimen data records, put the specimen ''dwc:occurrenceID'' in the ''ac:associatedSpecimenReference'' field in the image metadata CSV. | ||
*Use Audubon Core metadata, http://terms.tdwg.org/wiki/Audubon_Core_Term_List, with one record to go with each media record. The more you can flesh out the details of the image, the more likely it will be to be highly retrievable. | *Use Audubon Core metadata, http://terms.tdwg.org/wiki/Audubon_Core_Term_List, with one record to go with each media record. The more you can flesh out the details of the image, the more likely it will be to be highly retrievable. | ||
*Just like the ownership of catalog records, the media records need to be provided freely and with permission, and each record needs to have at least Creative Commons permission = "CC BY" | *Just like the ownership of catalog records, the media records need to be provided freely and with permission, and each record needs to have at least Creative Commons permission = "CC BY". | ||
==Error handling== | ==Error handling== | ||
Revision as of 17:29, 14 January 2014
Guidance to data providers when first considering sending their data to iDigBio
Audience: Data Providers, iDigBio data ingestion staff
This is the process description for
- the iDigBio staff to follow to assure that data are successfully and efficiently moved from data provider to the portal, available for searching.
- data providers to follow to assure that data are efficiently and accurately provided to the iDigBio staff.
Contact info
If you find yourself in need assistance, contact data@idigbio.org
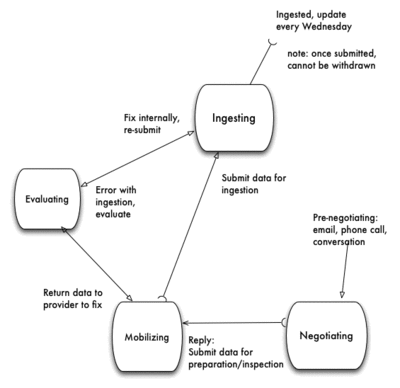
Process terminology
Processing steps, each step has a start and an end, signifying that it has moved to the next step.
- negotiating - in the process of determining provider's interest in data ingestion
- begins with email invitation to providers (in institutions, aggregators) to invite them to send their data to iDigbio specimen data portal
- open a Redmine ticket in category=Data Mobilizing
- ends with data exported by provider, ready for inspection and ingestion
- mobilizing - in the process of evaluating data being fit for ingestion
- begins with provider exported data and cursory inspection
- ends with data passing inspection and passing to ingesting state, Redmine ticket changes to category=Data
- ingesting - in the process of ingesting provider's data
- begins with Redmine ticket change to category=Data
- ends with
- data successfully ingested, ready for consumption
- report sent back to data mobilizing staff
- report sent to provider
- Redmine ticket set to Status= Closed
- evaluating - in the process of evaluating a failure to be ingested
- begins with ingestion failure
- evaluate ingestion failure, if data error - send it back to mobilizing state for corrections or
- evaluate ingestion failure, if ingestion error - make corrections
- ends with data re-submission to ingesting state
- begins with ingestion failure
Data requirements for data providers
Below are what we ask of the data to make it easily searchable in the cyberinfrastructure we provide.
There are 3 kinds of data files to submit for ingestion:
- specimen data with dataset metadata
- media data related to and attached by reference to specimen records with metadata (use of dwc:associatedMedia is not viewed as sending media)
- media files - e.g., non-archival .jpgs (see acceptable format here: https://www.idigbio.org/content/idigbio-image-file-format-requirements-and-recommendations)
Packaging for specimen data
In order of preference:
- DwC-A (Darwin Core Archive) in a RSS feed produced by IPT
- Custom DwC-A in an RSS feed produced by Symbiota
- Custom CSV or TXT (save the data in UTF-8 format to preserve diacritics in people and place names), option for only sending specimen data or only media data (DwC-A packaging required when sending both specimen and media data)
Sending data to iDigBio
- an RSS feed for ready access and update is our preference
- email the files to us
Specimen metadata
- Each specimen record should have a unique (within the dataset) identifier in the dwc:occurrenceID field. When the ingestion software detects duplicate identifiers, the duplicated records are flagged as an error and are not ingested. Identifiers, if not GUIDSs, are what is typically called the DwC (Darwin Core) triple:
<dwc:institutionCode> <dwc:collectionCode> <dwc:catalogNumber>. - If using a custom CSV, use field names that are as close to DwC terms as possible, additionally, make use of the MISC field names (local iDigbio extensions to DwC). The host association terms are an example of an extension found in the MISC. Use the XML style field names that include the domain of the schema, e.g., dwc:termName, ac:termName. Non-standard field names are not indexed and are not searchable.
Complete attribution and licensing
In order for each provider's data to be correctly attributed when found on the iDigBio portal, the following are important to complete:
- fill in your official institution code (dwc:InstitutionCode) (see and update http://GRBio.org)
- go to http://GRBio.org to get their coolID value for your institution to store in the dwc:institutionID and dwc:collectionID fields
- fill in the DwC global-to-the-dataset DwC record-level fields for intellectual property and licensing, e.g., dcterms:rightsHolder, dcterms:accessRights and dcterms:rights.
Permission to ingest
- the provider needs to have permission to submit their data
Data recommendations for optimal searchability
- put dates in ISO 8601 format, i.e., YYYY-MM-DD, e.g., 2014-06-22
- put elevation in METERS units in the elevation field without the units (e.g., the fields dwc:minimumElevationInMeters and dwc:maximumElevationInMeters already assume the numeric values are in meters, so there no need to include the units with the data)
- do not use unescaped newline characters
- do not export '0' in fields to represent no value, e.g., lat or lon
- make sure lat and lon coordinates are in decimal, and no N, S, E, W
- parse genus, species, infraspecific epithet if already aggregated into a scientific name
- include parsed higher taxonomy, at least kingdom and family if possible, and the intervening ranks if not controversial
Packaging for images/media objects
- Each media record should have a unique (within the dataset) identifier in the dwc:occurrenceID field.
- If submitting media records with specimen data records, put the specimen dwc:occurrenceID in the ac:associatedSpecimenReference field in the image metadata CSV.
- Use Audubon Core metadata, http://terms.tdwg.org/wiki/Audubon_Core_Term_List, with one record to go with each media record. The more you can flesh out the details of the image, the more likely it will be to be highly retrievable.
- Just like the ownership of catalog records, the media records need to be provided freely and with permission, and each record needs to have at least Creative Commons permission = "CC BY".
Error handling
When data are received from the provider during the mobilizing process step, they are evaluated for fitness. Once the evaluation is successful, the ingestion process moves from mobilizing to ingesting, and the data are submitted to the ingestion scripts. If an error condition occurs, the staff evaluate whether it is a script error or a data error. If it is the latter, the staff sends an email to the mobilizing staff who may contact the provider for changes. When the errors have been addressed, the mobilizing staff re-submit the data to the ingesting staff.
Use case for media and data
2 CSVs (scenario: one for image metadata, one for specimen data, plus uploaded images with > 1 image per specimen)
- Put specimen dwc:occurrenceID in the field ac:associatedSpecimenReference in the image metadata CSV (whose schema is in Audubon Core format)
- Generate a meta.xml file by hand and package up the files in a DwC-A like format. (No eml.xml required).
Sample scenarios of data transformations to prepare data for ingestion
- Example preparing specimen data from Illinois Natural History survey (INHS) fish collection from FileMakerPro
- Example of transformations on InvertNet image metadata dataset
Additional references
If you want to learn about acceptable Creative Commons licenses in iDigBio: